All published articles of this journal are available on ScienceDirect.
The Activity of α-glucosidase Inhibition of Pediococcus Acidilactici BAMA 4 Isolated from “Naniura” Traditional Foods from North Sumatera, Indonesia
Abstract
Background:
Type 2 diabetes is caused by unhealthy lifestyles, such as consuming foods rich in simple sugars and lack of exercise. One of the treatment therapies for this disease is α-glucosidase inhibitors. Some strains of the lactic acid bacteria (LAB) group typically exhibit α-glucosidase inhibitory activity, antimicrobial activity, acids and bile salt tolerance, and probiotic status.
Objective:
This study aims to isolate LAB from naniura, characterize and test its activity as an α-glucosidase inhibitor, as well as identify those with the highest activity.
Methods:
The method used to molecularly identify potentially good LAB was through the amplification of the 16S rRNA gene.
Results:
This study obtained a total of 9 strains using BAMA codes 1, 2, 3, 4, 5, 6, 7, 8, and 9. The cocci cell activity, gram-positive, and antibacterial activity of BAMA 4 against E.coli was 7.54 mm, while against S. aureus was 8.05 mm. The percentage of viability in acid and bile salt is 28.7% and 68.6%, respectively. Additionally, the proportion of α-glucosidase inhibition is 65%.
Discussion:
The BAMA 4 strain is a species of Pediococcus acidilactici. Pediococcus is a genus of gram-positive lactic acid bacteria in the Lactobacillacea family.
Conclusion:
BAMA 4 strain produced antibacterial optimally and its cells survived the growth in acid and bile conditions. The percentage of probiotics was relevantly high in this activity. This study will be helpful for other in-vivo research.
1. INTRODUCTION
Metabolic disorders are common pathologies, particularly polygenic diseases such as diabetes mellitus. The number of diabetic patients continues to grow globally, as the number of adult patients in the year 2017 increased to 8.8% of the world population and is expected to increase to 9.9% by the year 2045.
This shows that a population of 424.9 million people was affected with diabetes globally in 2017, whereas the number of patients is expected to increase to around 628.6 million people in 2045 [1]. The disease is considered to be the 7th leading cause of death. Furthermore, it is sometimes caused by the interaction of genetic and environmental factors, characterized by the absence of hormone secretion and resistance, causing disturbances in fat, protein, and carbohydrates [2].
Diabetes is classified into type 1 and type 2. Type 1 diabetes (T1DM) is a T-cell-mediated autoimmune disease that destroys insulin-producing pancreatic beta cells. The Type 2 counterpart (T2DM) is the most common diabetes characterized by insulin production and high blood glucose levels. α-glucosidase is an enzyme in the brush border membrane that catalyzes the digestion of carbohydrates. The inhibition of its activity has been shown to decrease glucose absorption and lower blood glucose levels. Therefore, α-glucosidase inhibitors are a therapeutic approach, and acarbose has been used as a potent inhibitor for diabetics. However, these drugs have side effects and secondary failure [3].
Probiotics are live microorganisms that confer a health benefit on the host when administered in adequate amounts. Probiotics are live microbial feed supplements that are beneficial and provide health effects for the host by improving its balance of microbial [4]. LAB is a group of bacteria with probiotic properties, and several reports have stated that some can inhibit glucosidase, hence, they are used to reduce the effects of diabetes [5]. LAB are classified as Generally Regarded As Safe (GRAS) organisms [4] and can be identified in some fermented foods, such as “naniura,” a local dish of North Sumatera made from fish and processed without cooking. According to a study, bacteria with antimicrobial activity can be isolated from naniura [6]. LAB that is isolated from food and known to have the ability to serve as as probiotics could be safely used as a drug source with no harmful effects to humans. Therefore, this research aims to isolate and screen LAB from naniura that exhibit α-glucosidase inhibitory activity. The species and strain of bacteria with the highest activity are then identified molecularly.
2. MATERIALS AND METHODS
2.1. Isolation, Characterization, and Catalase Assay of LAB
One gram of naniura from Medan restaurant, North Sumatera, was diluted in 100 mL de Man, Rogosa, and Sharpe (MRS) broth (Himedia, India) and incubated at 37°C for 24 h. The bacteria suspension was collected, the supernatant was pipetted at approximately 1 ml and then put into 9 ml sterile physiological NaCl solution (10-1). Subsequently, the dilution was performed up to 10-5, and after completion, 0.1 ml was pipetted from each dilution tube to MRS agar using a spreader, and then the Petri dishes were incubated for 48 h at 37°C. The growing bacteria were observed over the next 2 days.
Isolation of LAB was performed using MRS agar (Millipore, Merck, Germany), prepared by dissolving in distilled water, then sterilized in an autoclave, along with Petri dishes and other glassware. The sterile MRS agar was prepared in a Petri. The bacteria that grew were counted for the number of colonies. They were then purified by separating into several colonies and regrowing on MRS agar before reincubation. This purification was carried out until they became pure. Bacteria with the same colony shape and color in 1 Petri were considered single. Finally, the purified bacteria were stored in 25% (v/v0) glycerol at -20°C.
Gram staining was performed on about 9 isolates, and each strain was tested for gram color using a violet, iodine, 96% alcohol, and safranin solution. The test started by fixing the collected bacterial isolate with an ose needle and scratching it on a slide while being heated over a fire until the colonies dried up. The slide was dripped on the isolate with violet solution, left for 1 min, and rinsed with distilled water. Furthermore, it was dripped with iodine solution on the isolate, left for 1 min, and rinsed with distilled water. It was then rinsed with 96% alcohol until the color of the iodine and violet solution did not flow. The glass was again dripped with safranin solution on the isolate, left for 45 sec, then rinsed with distilled water. It was dried on paper towels, then observed under a microscope with a 100x lens. The gram color of the isolate is positive when it appears purple. However, when it is pink, then the gram color of the isolate is negative. Therefore, to classify a cell as bacilli, it should be rod-shaped, while a round cell is called cocci.
The catalase assay was conducted on all strains, the slide was prepared, and then some bacteria were scratched using an ose needle. H2O2 solution was dripped on the isolate and the bubbles formed were observed. The catalase assay was declared positive when bubbles were formed, implying that the strain can produce catalase enzyme. Meanwhile, it is declared negative when there is no formation of bubbles, which indicates that the strain cannot produce catalase enzyme.
2.2. Antibacterial Activity Assay
This test was conducted twice to test the ability of bacteria strains to inhibit the growth of pathogenic isolates, namely Escherisia coli and Staphylococcus aureus. At this stage, nine isolates were used because they had negative results in the catalase test. This test used chloramphenicol (30 µl) and MRS broth as positive and negative control, respectively. The test was performed using the diffusion method, starting with growing each strain and pathogen in MRS and Nutrient agar (NA) (Merck, Germany) in a test tube for 24 h. Each strain was collected using a needle ose part of the colony and then put into a physiological solution. Furthermore, the absorbance of the suspensions was calculated using UV-Vis spectrophotometry at a wavelength of 600 nm with an OD of 0.1. After observing that the number of cells followed OD 0.1, the suspense was ready for the assay. Each suspension of pathogens was collected with a sterile cotton swab and then streaked evenly onto the NA plate. After the pathogens were scratched onto the media, each assay strain was obtained by dipping sterile disk paper, draining, and placing on NA, which the pathogens had scratched. The media was incubated at 37°C for 24 h, and after 1 day, the inhibition zone was observed to be clear, after which the diameter was measured.
2.3. Acid and Bile Tolerance Assay
For the preparation of the stomach and intestines, the strains need to be identified to know their availability for each in acid and bile salt conditions because the strains were included as LAB from the previous assay. One of the strains with higher availability was taken for the next identification. MRS broth was adjusted to pH 3 with 1 M HCl. Bile solution was prepared by adding 0.3% bile acid factors (Jarrow Formula, Los Angeles) into MRS broth. Cell harvested by nine strains at an exponential growth phase (18 h) were inoculated in MRS broth, acid, and bile solution and incubated at 37°C for 4 h. The inoculates were adjusted to OD600 using spectrophotometry UV-Vis (Shimatzu). The following formula was used to measure the percentage of cells that survived in an acid solution [7]:
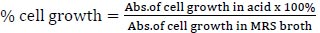 |
(1) |
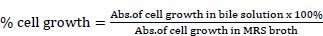 |
(2) |
2.4. α-Glucosidase Inhibition Assay
After incubation for 24 h in MRS broth, cultured BAMA 4 was harvested by centrifugation, and cells were disrupted using a homogenizer (Bactosoni, Bandelin, Germany). The disrupted cells were centrifuged at 4500 rpm for 15 min, and the supernatant was used for an inhibition assay, while acarbose was employed as a positive control. The determination of α-glucosidase was based on inhibiting crude glucosidase formation from the substrate sucrose [8]. This α-glucosidase inhibitory activity (%) was determined according to the following equation [9]:
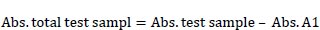 |
(3) |
Where: %test activity = Abs. total test sample/Abs. control x 100%. Total test sample absorbance = test sample absorbance + sucrose + crude enzyme. Absorbance A1= Absorbance of the test sample + sucrose.
2.5. Isolation DNA, 16S rRNA Amplification, and Sequencing of the Strain
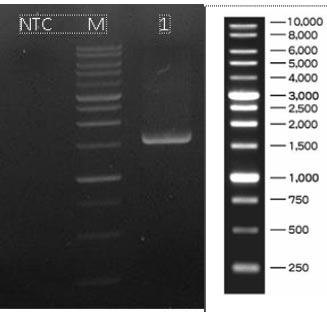
The isolation of DNA from the strain was conducted using the Quick-DNATM Bacterial Miniprep kit, and the procedure can be completed in 15 minutes. The strain was added directly to a ZR BashingBeadTM Lysis Tube (0.1 and 0.5 mm). Furthermore, it was rapidly and efficiently lysed by bead beating without using organic denaturants or proteinases. The DNA was then isolated and purified using Zymo-SpinTM Technology, which was ideal for downstream molecular-based applications, including PCR and array. The Genomic lysis buffer was added to a final dilution of beta-mercaptoethanol of 0.5% (v/v). Subsequently, 50-100 mg of bacterial cells resuspended in up to 200 μl of water or isotonic buffer (PBS) were placed in a ZR Bashing Bead™ lysis tube (0.1 mm and 0.5 mm). A total of 750 µl of BashingBead™ buffer was added to the tube. The ZR BashingBeadTM lysis tube (0.1 mm and 0.5 mm) was centrifuged in a microcentrifuge at 10,000xg for 1 min. Approximately 400 µl of the supernatant was transferred to a Zymo-SpinTM III-F filter. A total of 1200 µl of genomic lysis buffer was mixed with the filtrate in the collection tube from the previous step. The mixture at 800 µl was transferred to a Zymo-SpinTM IICR Column in a Collection tube and centrifuged at 10,000xg for 1 min. The flow-through was discarded from the Collection tube, and the precious step was repeated. A 200 µl DNA pre-wash buffer was added to the Zymo-SpinTM IICR Column in a new Collection tube and centrifuged at 10,000x g for 1 min. g-DNA wash buffer at 500 µl was mixed with the Zymo-SpinTM IICR Column and centrifuged at 10000x g for 1 min. The Zymo-SpinTM was transferred to a clean 1.5 ml microcentrifuge tube, and 100 µl DNA elution buffer was directly mixed with the column matrix. The mixture was centrifuged at 10000x g for 30 sec to elute the DNA. A strain designated primarily as LAB was identified in its 16S rRNA with 27F/1492R PCR primer. PCR amplification of genes was used (2x) My Taq HS Red Mix (Bioline, BIO-25048). Its products at 1 µl were assessed by electrophoresis with 0.8% TBE agarose at non-template control (NTC). The nucleotides were sequenced with Bi-directional. Meanwhile, the sequences were compared with those in the NCBI Genbank database (http://www.ncbi.nlm .nih.gov/BLAST/) using BLAST (NCBI, Bethesda, MD, USA).
3. RESULTS
3.1. Isolation, Characterization, and Catalase Assay of LAB
The total number of bacteria isolates obtained from this study was 9, with isolates codes as BAMA 1, 2, 3, 4, 5, 6, 7, 8, and 9. The characteristics of the isolates include convex shape, brownish-white color, small, and rounded edges. Gram staining and catalase test were carried out on isolates, and the following results are listed in Table 1.
| Isolate Codes | Cell Form | Gram Stain | Catalase Test | ||
|---|---|---|---|---|---|
| BAMA 1 | - | Coccus | - | Negative | - |
| BAMA 2 | - | Coccus | Positive | - | - |
| BAMA 3 | - | Coccus | - | Negative | - |
| BAMA 4 | - | Coccus | - | - | - |
| BAMA 5 | - | Coccus | - | Negative | - |
| BAMA 6 | - | Coccus | Positive | - | - |
| BAMA 7 | Bacil | - | - | Negative | - |
| BAMA 8 | - | Coccus | - | Negative | - |
| BAMA 9 | - | Coccus | Positive | - | - |
Based on the test results above, the nine isolates were negative in the catalase test. LAB is a bacterial that produces negative results in the catalase test. From the characteristics of LAB, which was negative from the catalase test, the nine isolates were considered LAB.
3.2. Antibacterial Assay
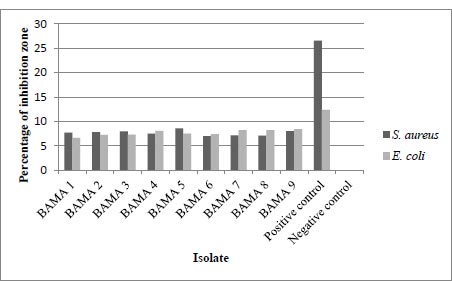
The results of the measurement of the diameter of the inhibition zone or the clear zone are in Table 2 below, also, the results of the average inhibition zone measurement include the size of the paper disc. This test was carried out twice, the diameter of the inhibition zone was measured vertically and horizontally. The positive control used in this assay was chloramphenicol 30 µg and the negative control was sterile MRS broth. The following are the results of the calculation of the average diameter of the antibacterial test from 9 isolates listed in Table 2 and shown in Fig. (1).
| Isolate Codes | Inhibition Zone Diameter against E. coli (mm) | Inhibition Zone Diameter against S. aureus (mm) |
|---|---|---|
| BAMA 1 | 7.73 | 6.65 |
| BAMA 2 | 7.82 | 7.27 |
| BAMA 3 | 7.95 | 7.31 |
| BAMA 4 | 7.54 | 8.05 |
| BAMA 5 | 8.58 | 7.51 |
| BAMA 6 | 6.99 | 7.42 |
| BAMA 7 | 7.14 | 8.23 |
| BAMA 8 | 7.11 | 8.22 |
| BAMA 9 | 8.03 | 8.43 |
| Positive control | 26.57 | 12.35 |
| Negative control | 0 | 0 |
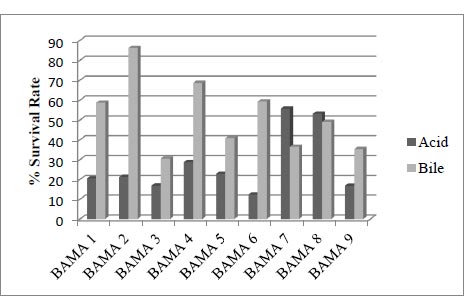
3.3. Acid and Bile Tolerance Assay
This compares the absorbance of cell growth in acid with normal media and the absorbance 600nm of cell growth in bile with normal media, with an incubation time of 4 hours.
3.4. Inhibition α-Glucosidase Activity
The α-glucosidase inhibition activity assay of the BAMA 4 strain was 65%, while that of acarbose was 70%. This indicates that the supernatant of isolate BAMA 4 grown in MRS broth media contained compounds that had the ability to inhibit the hydrolysis of crude glucosidase enzymes to sucrose.
3.5. Isolation DNA, 16S rRNA Amplification, and Sequencing of BAMA 4 Strain
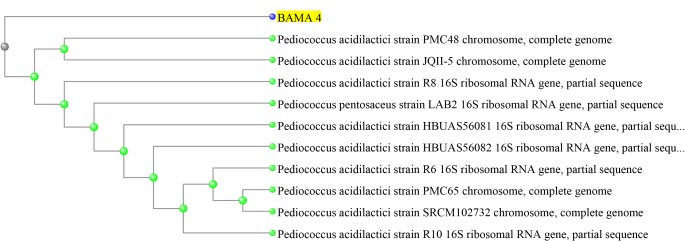
Based on the results of molecular identification, BAMA 4 strain was of Pediococcus acidilactici species, in the form of lactic acid bacteria, gram-positive and has cocci cells. Genomic DNA of P. acidilactici BAMA 4 (Fig. 2) was observed on Nanodrop methods and the absorbance can be seen in Table 3 (Fig. 3), while the sequences shown in Table 4 and results of electrophoresis PCR product (Fig. 4). The hit BLAST results against the NCBI database, excluding uncultured sample sequences and P. acidilactici and the phylogenetic tree can be seen in Fig. (5).

| Sample Name | Code Sample | Conc. (ng/µl) | A260/280 | A260/230 | Volume (µl) |
|---|---|---|---|---|---|
| BAMA 4 | 2430-1 | 62.4 | 1.91 | 0.17 | 30 |
Table 4.
| Sample Name | Sequences | ||||
|---|---|---|---|---|---|
| BAMA 4 |
Sequence Assembly 1465bp 1TCTGTCCACC TTAGACGGCT 61ACTCTCATGG TGTGACGGGC 121TGATCCGCGA TTACTAGCGA 181 CTGAGAACGG TTTTAAGAGA 241 ATTGTAGCAC GTGTGTAGCC 301 TTCCTCCGGT TTGTCACCGG 361 AATAAGGGTT GCGCTCGTTG 421 ACCATGCACC ACCTGTCATT 481 AGATGTCAAG ACCTGGTAAG 541CTTGTGCGGG CCCCCGTCAA 601 GATTACTTAA TGCGTTAGCT 661 ATCGTTTACG GCATGGACTA 721 CTCAGCGTCA GTTACAGACC 781 CGCATTTCCC CGCTACACAT 841 TCCAATGCAC TTCTTCGGTT 901 GCTCGCTTTA CGCCCAATAA 961 TGGCACGTAG TTAGCCGTGG 1021CACCCACGTT CTTCTTTAAC 1081GCGTTGCTCC ATCAGACTTG 1141AGTCTGGGCC GTGTCTCAGT 1201ATCGCCTTGG TGAGCCGTTA 1261TGATAGCAGA GCCATCTTTT 1321CATCAGTTTC CAGGTGTTAT 1381 GTCCGCCACT CACTTCGTGT 1441GGAAGTTCGT TCGACTTGCA |
AGCTCCTAAA GGTGTGTACA TTCCGACTTC TTAGCTAAAC CAGGTCATAA CAGTCTCACT CGGGACTTAA CTGTCCCCGA GTTCTTCGCG TTCTTTTGAG GCAGCACTGA CCAGGGTATC AGACAGCCGC GGAGTTCCAC GAGCCGAAGG ATCCGGATAA CTTTCTGGTT AACAGAGCTT CGTCCATTGT CCCAATGTGG CCTCCCCAAC AAAAGAAAAC CCCCTGCTTC TAAAATCTCA GTATA |
AGGTTACCCC AGGCCCGGGA GTGTAGGCGA CTCGCGGTTT GGGGCATGAT AGAGTGCCCA CCCAACATCT AGGGAACGCC TAGCTTCGAA TTTCAACCTT AGGGCGGAAA TAATCCTGTT CTTCGCCACT TGTCCTCTTC CTTTCACATT CGCTTGCCAC AAATACCGTC TACGAGCCGA GGAAGATTCC CCGATTACCC TAGGTAATGC CAGGCGGTTT TGGGCAGGTT TTCAGTGCAA | ACCGGCTTTG ACGTATTCAC GTTGCAGCCT CGCGACTCGT GATTTGACGT ACTGAATGCT CACGACACGA TAATCTCTTA TTAAACCACA GCGGTCGTAC CCCTCCAACA CGCTACCCAT GGTGTTCTTC TGCACTCAAG AGACTTAAAA CTACGTATTA ACTGGGTGAA AACCCTTCTT CTACTGCTGC TTTCAGGTCG GCCGCGGGTC TCTCTGTTAT ACNCCACGTG GCACGTCATA | GGTGTTACAA CGCGGCATGC ACAGTCCGAA TGTACCATCC CGTCCCCACC GGCAACTAGT GCTGACGACA GGTTGGCAGA TGCTCCACCG TCCCCAGGCG CTTAGTAATC GCTTTCGAGC CATATATCTA TCTCCCAGTT GACCGCCTGC CCGCGGCTGC CAGTTACTCT CACTCACGCG CTCCCGTAGG GCTACGCATC CATCCAGAAG ACGGTATTAG TTACTCACCC ATCAATTAAC |
4. DISCUSSION
This study successfully isolated 9 LAB strains with BAMA codes 1, 2, 3, 4, 5, 6, 7, 8, and 9, cell shape, and gram color, as shown in Table 1. In fermented foods, some bacteria can survive in pH 3, and strains of Lactobacillus, Weissella, and Leuconostac exist in kimchi [10]. LAB isolation obtained a strain of Lactobacillus fermentum (11). The catalase test of the 9 isolates was negative, indicating that LAB could not degrade H2O2 into H2O and O2. The strains obtained also produced a negative result [11]. LAB are probiotics used to improve the health of body, while those obtained from food survive using the nutrients in their host. Furthermore, they are bacteria living in acidic fermented foods.
Isolated LAB from the mango fruit (Mangifera indica L.) were M1 isolates [12]. The bacteria are also present in fermented milk drinks [13]. From the cell growth test results in acid and bile, the strain with more optimal survival was BAMA 4, with 28.7 and 68.6% percentage values in each condition. The value of bile was higher than that of 1, 2, 3, 5, 6, and 9 strains but lower than 7 and 8. Meanwhile, the percentage survival cells growth of the BAMA 4 strain in acid was higher than the value of 1, 3, 5, 6, 7, 8, and 9 strains (Fig. 2).
The ability to inhibit the growth of gram-positive bacteria E. coli and gram-negative bacteria S. aureus has been tested against isolates BAMA 1 – 9. From this activity, the BAMA 4 strain had more optimal ability for inhibiting E. coli and S. aureus than 8 other isolates (Table 2). Furthermore, the diameter of its inhibition zone was greater for S. aureus than for E. coli. Genes of E. coli were associated with main virulence factors such as adhesins, toxins with increased pathogenicity and caused human infection [14]. The resistance level of gram-negative bacteria was higher than its gram-positive counterpart [15]. This is because, in gram-negative cells, there is a permeability barrier on the outermost layer of the cell, such that antibacterial compounds are suitable to penetrate the cell and prevent binding to the intracellular side.
LAB is a probiotic that can inhibit pathogenic bacteria and form colonization in food as its host [16]. Similarly, LAB can kill pathogenic bacteria, especially gram-positive bacteria, by damaging cell membranes [17]. LAB can produce organic acid compounds in the form of fatty acids, hydrogen peroxide, and bacteriocins as antimicrobials [18]. Moreover, LAB have the ability to produce hydrogen peroxide, CO2, diacetyl, acetaldehyde, D-isomers of amino acids, reuterin, and bacteriocins [19]. Organic acid compounds produced by LAB can enter the cell membrane of pathogens, disrupt the transport system, and change the cell membrane's permeability. Furthermore, the acid compounds can be toxic in pathogenic cells. Acetic and lactic acid produced by LAB can interact with cell membranes causing the intracellular environment and protein to become acidic and denatured [17].
The 9 LAB strains were tested for their availability to survive in pH 3 and bile salt conditions. Microorganisms are declared as probiotics when they have characteristics such as originating from humans, not being pathogenic, being able to survive when passing through the intestines, being able to live in mucus, preventing pathogenic microbes, having a good effect on the immune system, and general health [20]. In other tests, the strains following BAMA 1, 2, 3, 4, 5, 6, 7, 8, and 9 have identified their availability to produce gas bubbles from the catalase test and their antibacterial activity. From screening these results, BAMA 4 isolates had a more optimal survival ability than the other 8 isolates. LAB isolated from dangke food, where the isolate could live in acidic media and the bacteria's cell cytoplasm is more alkaline [21]. This is because LAB can induce an Acid Tolerance Response (ATR) to respond to an acidic environment, form a hemostatic pH, and protect as well as improve the mechanism system in cells [22]. LAB can reduce the detergent effect by hydrolyzing bile salt using the hydrolase (BSH) enzyme, allowing LAB to survive in an alkaline environment under alkaline conditions (bile salt),. Furthermore, LAB can also increase glucose production as a source of nutrients from the bile salts for survival, using the available ATP.
The ability of the BAMA 4 strain to inhibit the degradation of sucrose into glucose by glucosidase is the ability of LAB in general, which has the potential to produce antidiabetic compounds, where in this study its inhibitory ability was 65% compounds. Ethanol extract from Swietenia mahagoni seed has availability as an inhibitor alpha-glucosidase with 18.647% for analysed hypoglycemia activity [23]. However, a study reported that acarbose has the availability inhibition of alpha-glucosidase with the value of 66-69% against alpha-amylase [24]. The percentage inhibition alpha-glucosidase from BAMA 4 included the best value for preventing T1DM and T2DM. Dat et al. [25] isolated five compounds from the crude extract of Bacillus sp. following: 2-(2-heptenyl)-3-methyl-4-quinolinone (9), 3-methyl-2-(2- nonenyl)-4-quinolinone (10), 2-phenylacetic acid (11), 4-hydroxybenzoic acid (12), and (−)-jasmonic acid. Triazoloquinazolines isolated from Saccharomyces cerevisiae type 1 showed alpha-glucosidase inhibition (AGI)’s activity with the highest value an IC50 83.87 µM [26]. LAB undergoes a fermentation process on polysaccharides in the intestine, producing a short-chain of fatty acids in the form of propionate and butyrate. These product compounds help the activity of G-protein coupled receptors (GPCRs) and glucagon-like peptides (GLP-1), which have the function of stimulating insulin secretion and reducing glucose in the body [27].
BAMA 4 strain was known to be a Pediococcus genus. Pediococcus is a genus of gram-positive lactic acid bacteria in the Lactobacillacea family. The genus Pediococcus consists of the species P. acidilactici, P. pentosaceus, P. damnosus, P. parvulus, P. inopinatus, P. halophilus, P. dextrinicus, and P. urinaeequi. Based on the results of sequencing, BAMA 4 isolates were P. acidilactici species. P. acidilactici is reported to have the ability to produce bacteriocins, which are proteins that can inhibit pathogens [28]. The genus Pediococcus is a facultative, non-motile, and non-spore-forming anaerobic bacterium [29]. The ability of P. acidilactici as a probiotic has the ability to live in an acidic and alkaline environment, is able to produce bacteriocins, and has antidiabetic compounds against mice simulated with diabetes [30].
CONCLUSION
A total of 9 LAB strains were obtained, with codes following: BAMA 1, 2, 3, 4, 5, 6, 7, 8, and 9. BAMA 4 strain most optimally produced antibacterial and its cells more survived growth in acid and bile conditions. Species of BAMA 4 strain as P. acidilactici. Inhibitor-producing P. acidilactici BAMA 15 value is 65% while acarbose is 70%. The percentage of probiotics was relevantly high in this activity.This study can be useful for further research in-vivo.
AUTHORS’ CONTRIBUTIONS
D.R and E.G contributed to conceptualization, methodology, software, draft preparation, and editing. E.F and F.P contributed to conceptualization, data curation, and draft preparation. C.N.-G and A.N.-N contributed to methodology, draft preparation, and editing. All authors have read and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| T2DM | = Type 2 counterpart |
| GRAS | = Generally Regarded as safe |
| ATR | = Acid tolerance response |
| BSH | = Bile salt hydrolase |
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
FUNDING
The authors are grateful to The Directorate of Research, Technology, and Community Service (DRTPM) for financing this study. Also, Research, Technology, and Higher Education of the Republic of Indonesia 2022 with contract number: 63/LL1/LT/K/2022, under the scheme Research of Doctoral Dissertation (PDD), awarded to Chrismis Novalinda Ginting as the principal investigator.
ACKNOWLEDGEMENTS
The authors are grateful to the Department of Pharmacy and Doctoral Program of Medicine, Faculty of Medicine, Prima Indonesia University for their kind support in the present research work. The authors are grateful to The Directorate of Research, Technology, and Community Service (DRTPM) for financing this study. Also, Research, Technology, and Higher Education of the Republic of Indonesia 2022 with contract number: 63/LL1/LT/K/2022, under the scheme Research of Doctoral Dissertation (PDD), awarded to Chrismis Novalinda Ginting as the principal investigator.


