Expression and Enzyme Kinetics of Fused Plasmodium falciparum Orotate Phosphoribosyltransferase and Orotidine 5′-monophosphate Decarboxylase in Different Escherichia Coli
Abstract
Background:
Fusion of the last two enzymes in the pyrimidine biosynthetic pathway in the inverse order by having COOH-terminal orotate phosphoribosyltransferase (OPRT) and NH2-terminal orotidine 5′-monophosphate decarboxylase (OMPDC), as OPRT-OMPDC, has been described in many organisms.
Objective:
The study aimed to select the optimum host cell and temperature for expressing the recombinant fusion OMPDC-OPRT having the enzymatic activity.
Methods:
We constructed gene fusions of the human malaria parasite Plasmodium falciparum OMPDC-OPRT (1,836 bp) in the pTrcHisA vector and expressed it as a 6xHis-tag bifunctional protein in three Escherichia coli strains (BL21(DE3), TOP10, Rosetta) at 18°C and 25°C. The recombinant bifunctional protein was partially purified by Ni-nitrilotriacetic acid affinity chromatography and confirmed via Western blot and LC-MS/MS. The enzyme kinetics of OPRT and OMPDC was assessed.
Results:
Specific enzymatic activities of both OPRT and OMPDC domains expressed in E. coli BL21(DE3) cells were approximately eight-to-nine-fold higher than those in the TOP10 cells at 18°C. However, the specific activities of both domains expressed in the TOP10 cells were twice higher than those of the BL21(DE3) cells at 25°C. Very low and no enzymatic activities were observed when the constructed vector was expressed in the Rosetta cells at both induction temperatures. The bifunctional enzyme had specific activities of the OPRT and OMPDC domains in a ratio of 1:2. Kinetic study values of the OPRT domain in the bifunctional OMPDC-OPRT enzyme were found to be relatively low at µM level and at the perfect catalytic efficiency (kcat/Km).
Conclusion:
The recombinant fusion of OMPDC-OPRT exhibited a high expression level of E. coli BL21(DE3) at 18°C. The kinetic parameter is greater than 108M-1s-1.
1. INTRODUCTION
Malaria fever is an endemic disease known to be caused by Plasmodium species. In particular, P. falciparum is a major parasite found in Africa, whereas P. vivax is identified as the predominant parasite outside of Africa [1]. Plasmodium species have a different pyrimidine nucleotide synthesis compared to human and other mammalian cells, which cannot salvage preformed pyrimidine bases or nucleosides from the human host, except that P. falciparum exhibited minor pyrimidine synthesis by salvage pathway. It relies solely on nucleotides synthesized through the de novo pathway [2].
The de novo pathway involves six sequential enzymes catalyzing the conversion of the following precursors, i.e., HCO¬3-, ATP, Glu, Asp, and 5-phosphoribosyl-1-pyrophos-phate (PRPP), to form uridine 5′-monophosphate (UMP). The last two steps of UMP synthesis require the addition of ribose 5-phosphate from PRPP to orotate through orotate phosphoribosyltransferase (EC 2.4.2.10, OPRT) to form orotidine 5′-monophosphate (OMP) and the subsequent decarboxylation of OMP to form UMP through orotidine 5′-monophosphate decarboxylase (EC 4.1.1.23, OMPDC). Both enzymes are expressed and act as a bifunctional enzyme complex in the form of heterotetrameric (PfOPRT)2 (PfOMPDC)2, but the genes are separated on chromosomes 5 and 10, respectively. In humans, both enzymes are transcripts from chromosome 3, existing as a bifunctional UMP synthase (UMPS), OPRT at the N-terminus, and OMPDC at the C-terminus. In contrast, they are expressed separately as monofunctional enzymes in bacteria. Interestingly, OPRT and OMPDC from Leishmania donovani and Trypanosoma cruzi are the inverse order of OMPDC-OPRT, which, through evolution, had shown lateral gene transfer, split, and finally, a refusion of the gene [3]. Plasmodium evolution is similar to Leishmania donovani and Trypanosoma cruzi, with an OPRT and OMPDC gene fusion possibly occurring in an inverse order.
The possibility of a fusion of the P. falciparum OMPDC-OPRT gene has been reported. There have been reports on rearrangements of protein domains like closely related species: Leishmania donovani and Trypanosoma cruzi. So, we had to construct a gene in the fusion of the OMPDC-OPRT gene in different E. coli host cells to obtain the high-performance expression protein for examining enzyme kinetics and choose the optimum host cell for the recombinant OMPDC-OPRT fusion enzyme expression.
2. MATERIALS AND METHODS
2.1. Construction of Plasmid
PCR technique was used to amplify the OMPDC gene (969 bp), the forward primer 5′CGGGATCCGCCATGGGTTTT-AAGGTAAAATTAG3′, and the reverse primer 5′GCCTCG-AGCGATTCCATATTTTGCTTTAAG3′; the primers were then designed to introduce the restrict site for BamHI and XhoI, respectively. OPRT gene (843 bp) was amplified by PCR using the forward primer 5′GCCTCGAGATGACGACGAT-AAAAGAGAATG3′ and the reverse primer 5′GCCTGCA-GCGCTCATATCATCGACTGTATATCGTC3′, which intro-duced XhoI and PstI restriction sites, respectively (supplement 1). Each PCR product was ligated separately into the Zero Blunt TOPO cloning vector (Invitrogen), as previously described [4, 5]. The OMPDC gene was first cloned into a pTrcHisA expression vector (Invitrogen), and then subsequently sub-cloned with the OPRT gene, producing a protein fused to 6xHis-tag at the N-terminus (Fig. 1). Nucleotide sequencing of the construct OMPDC-OPRT gene was determined [6]. The constructed plasmid was then transformed into competent E. coli BL21(DE3) (Promega), TOP10 (Invitrogen), and Rosetta (Novagen) cells (Table 1).

| E. coli Strain | Genotype | Source or Reference |
| BL21 (DE3) | F-, ompT, hsdSB (rB-, mB-), dcm, gal, λ(DE3), pLysS, Cmr | Promega |
|
TOP10 |
F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK λ- rpsL(StrR) endA1 nupG | Invitrogen |
| Rosetta | F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE (CamR) | Novagen |
2.2. Recombinant Protein Expression and Purification
The E. coli cells were grown in LB medium at 37°C to an OD600 nm of 0.4-0.6 and induced with 1 mM isopropyl α-D-thiogalactopyranoside (IPTG) at 18°C for 18 h and 25°C for 18 h, harvested by using a refrigerated incubating shaker (8000×g), and stored at −80°C as a cell paste until use [5]. The cell paste was processed to complete lysis by using a lysis buffer containing 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 1% Triton X-100, 0.1%(w/v) lysozyme, 0.1% RNase, and protease inhibitor cocktail (Boehringer Mannheim), and then sonicated by ultrasonic SonopulsTM homogenizer (Bandelin). The crude extract was centrifuged at 10,000 x g at 4°C for 30 min. The supernatant fluid was loaded onto nickel-nitrilotriacetic acid agarose affinity chromatography (Ni-NTA, Qiagen) to purify the recombinant protein using 6xHis-tag at the N-terminus. The Ni-NTA column (1 ml bed volume) was equilibrated with buffer A (50 mM Tris-HCl, pH 8.0; 300 mM NaCl; 10 mM imidazole). The column was washed twice with 5 ml buffer A and twice with 5 ml buffer B (50 mM Tris-HCl, pH 8.0; 300 mM NaCl; 20 mM imidazole). The recombinant protein was eluted with 6 ml of buffer C (50 mM Tris-HCl, pH 8.0; 300 mM NaCl; 250 mM imidazole) [5]. The protein was further purified by Hi-Trap Q anion-exchange chromatography (GE Healthcare), equilibrated with 50 mM Tris-HCl pH 8.0, and washed with 50 mM Tris-HCl pH 8.0; 100 mM NaCl, 50 mM Tris-HCl pH 8.0; 250 mM NaCl and 50 mM Tris-HCl pH 8.0; 300 mM NaCl. The recombinant protein was eluted with 50 mM Tris-HCl pH 8.0; 500 mM NaCl.
2.3. Enzymatic Activity Assay and Kinetic Studies
OPRT and OMPDC activities of the recombinant OMPDC-OPRT protein were measured at 37°C in a quartz cuvette using a UV-visible Shimadzu spectrophotometer model UV 1601 with a temperature control device. The OPRT reaction mixture (1 ml) contained 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 250 µM DTT, 250 µM orotate, and 5-50 µl enzyme, and was incubated at 37°C for 1 min. The reaction was started by adding 250 µM PRPP, monitoring the decrease in absorbance of the substrates orotate at 295 nm for up to 3 min. For the OMPDC assay, the reaction mixture (1 ml) contained 50 mM Tris-HCl, pH 8.0, 250 µM DTT. The enzyme (5-50 µl) was incubated at 37°C for 1 min; the reaction was then started by adding 250 µM OMP, which was followed by a linear progression curve of absorbance change at 285 nm up to 3-6 min [4, 5]. The kinetic parameter, e.g., Km, Vmax and kcat of OPRT (>90% purity), was determined by varying PRPP concentrations from 12.5 µM to 250 µM [4, 5, 7].
2.4. Protein Assay
Protein concentrations were determined using the Bradford assay using bovine serum albumin as standard [8].
2.5. SDS-PAGE and Western Blot
SDS-PAGE was performed on a BioRad minislab gel apparatus with a 5% acrylamide stacking gel and 10% acrylamide running gel in the discontinuous buffer system of Laemmli [9]. The gels were stained with Coomassie Brilliant Blue R-250 dye. Western blot was analyzed to confirm the 6xHis-tag recombinant protein having PfMPDC-OPRT, which was detected using a monoclonal antibody directed against 6xHis-tag, as described [10].
2.6. In-gel Digestion and Liquid Chromatography-mass Spectrometry and Proteomic Data Analysis
The recombinant OMPDC-OPRT protein on SDS-PAGE was cut into small pieces and then destained and digested into small peptides by trypsin. The tryptic peptides were processed with a NanoAcquity system (Waters Corp.), and then tryptic peptides were analyzed using an SYNAPTTM HDMS mass spectrometer (Waters Corp.). LC-MS/MS data were analyzed by using MassLynx version 2.3 software, as described [11].
3. RESULTS
3.1. Functional Expression in Different E. Coli Host Cells
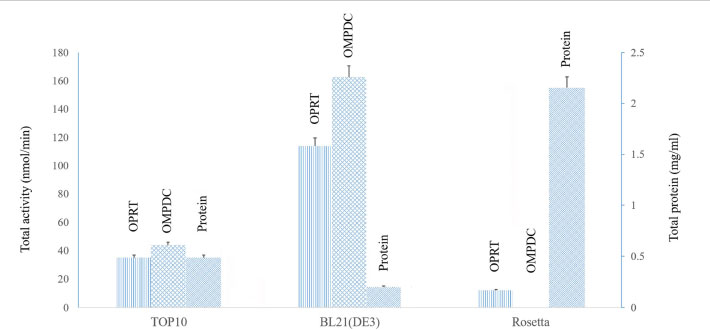
In this study, the artificial P. falciparum OMPDC-OPRT fusion gene containing 1,836 base pairs was produced from similarly close related species during evolution by having an amino acid identity of 31% and ligated into pTrcHisA expression plasmid. The constructed plasmid was transformed into three different E. coli host cells (TOP10, BL21(DE3), and Rosetta). The recombinant bifunctional OMPDC-OPRT protein was expressed under 1 mM IPTG induction at 18°C for 18 h.
| E. coli | 18 °C | 25 °C | ||||
|
Protein (mg) |
OPRT (nmol/min)a |
OMPDC (nmol/min)a |
Protein (mg) |
OPRT (nmol/min)a |
OMPDC (nmol/min)a |
|
| TOP10 | 0.4870 | 35.04 | 43.78 | 0.3697 | 70.12 | 97.24 |
| BL21(DE3) | 0.2023 | 113.91 | 162.59 | 0.8486 | 109.40 | 106.57 |
| Rosetta | 2.1543 | 12.08 | 0 | 1.1877 | 6.9 | 0 |
From 1 L of the cultures, the cell paste containing the recombinant bifunctional OMPDC-OPRT from E. coli TOP10, BL21(DE3), and Rosetta cells was 7.0 g, 4.3 g, and 5.0 g wet weight, respectively. The cell paste was lysed as described above; the soluble protein was then obtained after centrifugation. It was purified by Ni-NTA affinity chromatography. The bifunctional OMPDC-OPRT enzyme was eluted at 250 mM imidazole. Each of the bifunctional enzymes was then assayed separately. The total activity of OPRT and OMPDC from E. coli BL21 (DE3) was higher than that of TOP10 and Rosetta host cells, respectively. However, the total protein of bifunctional OMPDC-OPRT was the highest when expressed in Rosetta cells; however, relatively low and no OPRT and OMPDC activities were achieved, respectively (Fig. 2 and Table 2).
3.2. Functional Expression at Different Temperatures
The recombinant bifunctional OMPDC-OPRT enzyme was expressed at 18°C and 25°C, compared to E. coli TOP10, BL21 (DE3), and Rosetta host cells. The recombinant bifunctional OMPDC-OPRT enzyme was purified from the soluble protein fraction of the cell paste by Ni-NTA affinity chromatography. At 18°C, specific enzymatic activities of both OPRT and OMPDC domains expressed in E. coli BL21(DE3) cells (OPRT = 563.07 + 34.3 nmol/min/mg protein, OMPDC = 803.70 + 13.4 nmol/min/mg protein, n=4) were approximately eight- to nine-fold higher than those in the TOP10 cells (OPRT = 71.95 + 2.7 nmol/min/mg protein, OMPDC = 89.90 + 10.4 nmol/min/mg protein, n=4). However, the specific activities of both domains expressed in the TOP10 cells (OPRT= 189.67 + 17.5 nmol/min/mg protein, OMPDC = 263.02 + 16.9 nmol/min/mg protein, n=4) were two-fold higher than those of the BL21(DE3) cells (OPRT= 128.92 + 19.3 nmol/min/mg protein, OMPDC 125.58 + 45.1 nmol/min/mg protein, n=4) cultivated at 25°C. There were very low and no enzymatic activities when the constructed vector was expressed in the Rosetta cells at both temperatures (Table 2). Thus, the bifunctional OMPDC-OPRT enzyme had slightly different specific activities of OPRT and OMPDC domains with a ratio of approximately 1:1.
3.3. SDS-PAGE and LC-MS/MS of Recombinant Bifunctional OMPDC-OPRT Enzyme
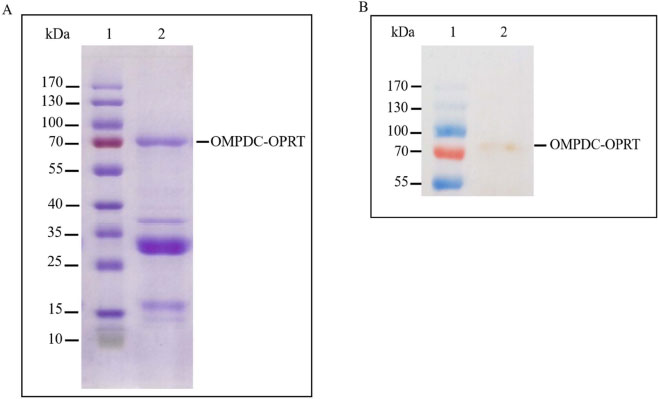
The recombinant protein was checked with a protein band on SDS-PAGE; as per the findings, the protein band was found to be highly expressed from Rosetta, TOP10, and BL21 (DE3), respectively. SDS-PAGE examined the recombinant bifunctional OMPDC-OPRT enzyme; the molecular mass of OMPDC-OPRT protein was 68+4 kDa (n = 4), corresponding to the predicted molecular mass of 70,812 Da from the deduced amino acid sequence of the protein (Fig. 3). The recombinant protein was confirmed by Western blot; the expected protein band from Rosetta was not shown.
The amino acid sequence of the enzyme was confirmed using LC-MS/MS after trypsin in-gel digestion of the bifunctional protein at 70 kDa band from the SDS-PAGE gel and proteomic analysis to identify the following sequences, that is, 476-KYSNIFYLYDRK-487, 554-KVVAFIVLLNRN -565, and 585-RVGIPLYSILSYKD-598, corresponding to the amino acid sequence in the crystal structure of P. falciparum OPRT (ID: gi|529281050) (Table 3).
| Peptide a | Peptide Score b | MH+ (Da) c |
| K.VVAFIVLLNR.N | 29 | 1142.72 |
| R.VGIPLYSILSYK.D | 39 | 1351.78 |
| K.YSNIFYLYDR.K | 28 | 1352.64 |
b The results were tested by the Decyder MS program with significance at p<0.05, whereas peptide score is the score of each peptide identification, high peptide score indicates high significance.
c MH+ (Da) is a peptide mass.
3.4. Purification of Recombinant Bifunctional OMPDC-OPRT from E. coli BL21(DE3) Expression at 18°C
The recombinant bifunctional OMPDC-OPRT enzyme from E. coli BL21(DE3) was purified by two sequential steps of Ni-NTA affinity and Hi-Trap Q anion-exchange chromatography. The recombinant OMPDC-OPRT enzyme was eluted with buffer C having 250 mM imidazole from the Ni-NTA affinity chromatography, which had a specific activity of OMPDC and OPRT as 803.72 and 563.08 nmol/min/mg protein, respectively. At the Hi-Trap Q anion-exchange chromatography, the enzyme was eluted by 50 mM Tris-HCl, pH 8.0, and 500 mM NaCl. The bifunctional OMPDC-OPRT enzyme used the high salt elution, which was different from the low salt elution of monofunctional OPRT and OMPDC [4, 7, 12-16]. The purity recombinant enzyme exhibited a specific activity of OMPDC with 631.7 nmol/min/mg protein and OPRT with 578.6 nmol/min/mg protein; the stoichiometry of OMPDC and OPRT was about 1:1 (Table 4). The results of both enzyme activities at the Hi-Trap Q anion-exchange chromatography recovered 72.2% yield for OPRT and 55.2% yield for OMPDC, compared to the Ni-NTA affinity chromatographic step. The purity of the recombinant bifunctional protein was analyzed using SDS-PAGE, of which its molecular mass of monomeric OMPDC-OPRT was 68+4 kDa (n = 4).
| Step |
Total Protein (mg) |
OPRT | OMPDC | ||
| Activity (nmol/min) |
Specific Activity (nmol/min/mg) |
Activity (nmol/min) |
Specific Activity (nmol/min/mg) |
||
| Ni-NTA | 0.2023 | 113.91 | 563.08 | 162.59 | 803.72 |
| Hi-Trap Q | 0.1421 | 82.22 | 578.63 | 89.77 | 631.74 |
3.5. Kinetic Parameters
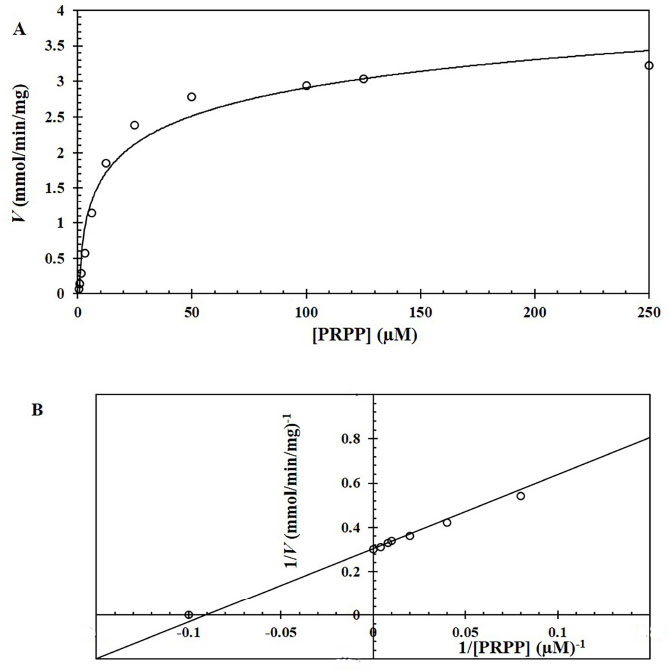
The Km and Vmax values of the OPRT domain in the bifunctional OMPDC-OPRT enzyme expressed from E. coli strain BL21 were determined using various concentrations of substrate PRPP. Kinetic parameters of the OPRT in the bifunctional enzyme were determined as Km= 9.3 + 0.5 µM, Vmax = 2,994 µM/min/mg protein, kcat = 3,534 s−1, and kcat/Km = 3.8 x 108 M−−1s−1. The Km of OPRT was relatively low at µM level, and the perfect catalytic efficiency (kcat/Km) was achieved with a valve greater than 108 M−1s−1 (Fig. 4). Another part of the enzyme, the OMPDC domain was determined at Km = 2.0 + 0.1 µM, Vmax = 835 µM/min/mg protein, kcat = 986 s−1, and kcat/Km = 4.9 x 108 M−−1s−1.
4. DISCUSSION
P. falciparum has been identified as one of the Plasmodium species responsible for most deaths from malaria fever in humans. OPRT and OMPDC are enzymes at the last two steps in the pyrimidine biosynthetic pathway to generate UMP from orotate and PRPP. The PfOPRT and PfOMPDC genes are encoded separately and have unique insertion genes that play a role in protein binding between OPRT and OMPDC, and function in the multienzyme complex as (OPRT)2(OMPDC)2 [4, 11, 15, 16]. In humans, OPRT and OMPDC are encoded from one gene that expresses OPRT at the N-terminus and OMPDC at the C-terminus as UMPS. The inversely linked OMPDC-OPRT gene fusion was present in the parasitic kinetoplastids (Leishmania donovani and Trypanosoma cruzi). In most prokaryotes and lower eukaryotes, they exist as monofunctional proteins [12-14]. So, the PfOMPDC-OPRT fusion gene is useful for studying gene evolution and function. The PfOPRT and PfOMPDC genes contain high A+T insert in DNA while never being found in the E. coli gene [17]. The Plasmodium protein expression in the E. coli system reported the correct protein that did not have codon bias. The previous study of monofunctional OPRT and monofunctional OMPDC demonstrates the correct amino acid sequence [4].
Previously, we had constructed the PfOMPDC-OPRT fusion gene and ligated it into the pTrcHisA plasmid [6]. Data analysis of the amino acid sequence of OMPDC-OPRT using the Clustal Omega program showed ~29.86% identity between PfOMPDC-OPRT and L. donovani. The amino acid sequence was compared as nearly phylogenetic [3]. The P. falciparum OPRT and OMPDC were noted to have an external and internal insert amino acid that shows protein-protein interaction in the active form of the enzyme between two molecules of OPRT and two molecules of OMPDC [5, 11].
The recombinant plasmid was transformed into three different E. coli host cells, that is, BL21 (DE3), Rosetta, and TOP10. The cells are different in terms of promoter and codon usage. The host cells were compared for protein expression and activity performance. The previous study showed the construction and expression of the recombinant protein of PfOMPDC, PfOPRT, and other proteins from Plasmodium to have identical amino acid sequences from that of P. falciparum 3D7 [18]. The expression of separate OPRT and OMPDC in E.coli TOP10 was observed, and verification of amino acid sequence and confirmation by activity test reported high activity of the same enzyme from P. falciparum and PfDHOase expressed from E. coli BL21 (DE3) [19]. In this study, the recombinant protein was expressed at 18°C and 25°C in the different host cells, that is, BL21 (DE3), TOP 10, and Rosetta. So, the recombinant protein was checked with a protein band on SDS-PAGE; as per the findings, it was found that protein was highly expressed from Rosetta, TOP10, and BL21 (DE3). The recombinant protein was confirmed by Western blot; the expected protein band from Rosetta was not shown. Recombinant protein from BL21 (DE3) and TOP 10 demonstrated activity, but the recombinant protein from Rosetta had no activity under the same expression conditions. The E. coli Rosetta strain is a BL21 derivative designed to increase the expression of eukaryotic proteins that contain codons rarely used in E. coli.
Protein expression can be affected by temperature; E. coli strain BL21 (DE3) and TOP10 protein were expressed at 18°C and 25°C, and the cell pastes from different temperature doses were deemed not significant. The activity of recombinant enzyme expression at 18°C was higher than the expression at 25°C. So, when we compared the activity between the different host cells at the same temperature, it was shown that at 18°C, the activity of recombinant protein from E. coli BL21 (DE3) was eight- to nine-fold higher than the recombinant protein from E. coli TOP10. But at 25°C, the activity of recombinant protein from E. coli TOP10 was shown to be two-fold higher than that in E. coli BL21 (DE3). The temperature was noted to affect the protein expression. While the high temperature was used as the expression condition for the expression protein, the expression results showed protein molecules in inclusion bodies. The optimal temperature for the expression system was the same as that of the monofunctional constructs at 18°C [3, 4].
The data of enzyme kinetics were same for bifunctional enzymes expressed from E. coli strain TOP10 in our previous study [6]. The kinetic parameter of the OPRT domain of the bifunctional enzyme, a study of how the PRPP concentration varies from 12.5 µM to 250 µM and the orotate is fixed at the saturated concentration, showed a 102- to 103-fold higher catalytic efficiency than OPRT monofunctional enzyme, indicating that the bifunctional enzyme expresses perfectly efficient catalysis and low µM level about three- to seven-fold of both domains. The kinetics of PfOPRT and PfOMPDC reported to be active in monofunctional form, multienzyme complex, and bifunctional form [4, 6, 7]. On the other hand, the structures of L. donovani OPRT and OMPDC were expressed from one gene as a bifunctional enzyme, which showed the enzyme kinetics as LdUMPS, but no function of OPRT was observed when it was expressed as a monofunctional enzyme and multienzyme complex: interaction between dimer of monofunctional OPRT and dimer of monofunctional OMPDC [20].
CONCLUSION
The fusion gene of OMPDC-OPRT has been found in many organisms, ranging from cyanobacteria to protozoa parasites, including the Plasmodium phylum. So, we constructed gene fusions in an inverted order, with the sixth enzyme at the N-terminus and the fifth enzyme at the C-terminus, which was likely to be phylogenetically close to L. donovani. The recombinant bifunctional protein OMPDC-OPRT of P. falciparum was expressed with high efficiency by E. coli BL21(DE3) at 18°C. The kinetic parameter was greater than 108 M−1s−1.
LIST OF ABBREVIATIONS
| OPRT | = Orotate phosphoribosyltransferase |
| OMPDC | = Orotidine 5′-monophosphate decarboxylase |
| PRPP | = Phosphoribosyl-1-pyrophosphate |
| UMP | = Uridine 5′-monophosphate |
| OMP | = Orotidine 5′-monophosphate |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No humans or animals were used for the studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the ResearchGate at https://www.researchgate.net/profile /Waranya-Imprasittichai/research.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors thank the Navamindradhiraj University for supporting the research. The authors thank the Faculty of Medicine, Vajira Hospital, for the grant provided for the proofreading and editing of this manuscript.


