All published articles of this journal are available on ScienceDirect.
Study of the Immunogenicity of Combination of Recombinant RBD (Omicron) and Nucleocapsid Proteins of SARS-CoV-2 Expressed in Pichia Pastoris
Abstract
Background:
SARS-CoV-2 is a virus responsible for the COVID-19 pandemic that began in late 2019. This pandemic has had a devastating impact worldwide, resulting in over 6.95 million deaths. The development of effective vaccines against the virus is crucial for preventing infection and reducing the severity of the disease. Objective: This study aimed to obtain the recombinant receptor-binding domain (RBD) and the nucleocapsid (N) proteins of SARS-CoV-2 as well as assess the immunogenicity of the combination of these recombinant proteins.
Methods:
The recombinant plasmids encoding the receptor-binding domain (RBD) of the spike protein of the Omicron variant and the nucleocapsid protein of SARS-CoV-2 were cloned into the yeast Pichia pastoris. The optimal fermentation conditions were established for recombinant P. pastoris strains. The methods for the isolation and purification of the target recombinant RBD and nucleocapsid proteins were developed. The immunogenicity of the purified recombinant proteins was evaluated by injecting them into mice and analyzing the specific IgG antibody responses using ELISA.
Results:
The study found that RBD and N proteins, as well as their combination, showed antigenic specificity and were highly immunogenic in mice. The immunogenicity was measured by determining the antibody titer, which represents the concentration of antibodies produced in response to the antigen. The antibody titers were 1:60000 for both RBD and N proteins, and 1:80000 for their combination.
Conclusion:
These findings suggest that the expressed proteins could be potential candidates for the development of vaccines or immunological diagnostic test kits for combatting or detecting the Omicron variant of SARS-CoV-2.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as the virus causing COVID-19 infection, is a contagious respiratory illness that emerged in late 2019 and quickly became a global pandemic. It is an RNA virus with a single-stranded positive RNA structure that leads to the development of COVID-19, an acute respiratory infection [1, 2]. By the end of 2022, researchers had developed over 230 potential vaccines using different methods to combat and prevent COVID-19 [3]. SARS-CoV-2 continues to undergo rapid mutations, leading to the emergence of dangerous variants that possess traits, such as increased transmissibility and the ability to evade neutralizing antibodies that have been previously effective against known strains. As a result, some of the vaccines that have been previously developed have shown reduced effectiveness against these new variants. This situation has contributed to the widespread transmission of the infection on a global scale [4-7]. Currently used vaccines based on protein subunits mainly target the receptor-binding domain (RBD) of the surface S (spike) protein of the virus, which has a high affinity for human angiotensin-converting enzyme 2 (ACE2) receptor and uses it as an entrance point [8-13]. Indeed, the spike protein of SARS-CoV-2, which is a crucial target for current vaccines, has been found to be subjected to a significant number of mutations. Notably, the Omicron variant (SARS-CoV-2 B.1.1.529), which emerged as a widespread strain, exhibited more than 30 mutations in its spike protein [14-17]. These mutations have raised concerns among health experts and researchers because they have the potential to impact various aspects of the virus' behaviour, including transmissibility, immune evasion, and vaccine effectiveness [18]. In this regard, the SARS-CoV-2 nucleocapsid (N) or its combination with the S protein may be the most suitable candidate for the development of effective vaccines. The N protein of SARS-CoV-2 is highly conserved and shows relatively little variation across different viral variants. In contrast, the spike protein of SARS-CoV-2 tends to accumulate a higher number of mutations. This is primarily because the spike protein plays a crucial role in facilitating viral entry into cells and interacting with the human immune system. As a result, mutations in the spike protein can have significant implications for the virus' infectivity, transmissibility, and evasion of the immune response [19-23]. Furthermore, the N protein of SARS-CoV-2 is highly immunogenic, meaning it triggers a strong immune response in the body. During infection, the N protein is expressed in significant amounts, resulting in a high titer of IgG antibodies specific to the N protein being present in the serum. This immune response to the N protein reflects its importance as a potential target for the development of vaccines and diagnostic tests against COVID-19 [24, 25]. Some antigenic regions of the N protein of SARS-CoV-2 are recognized by T cells on the surface of infected cells, which indicates the role of the N protein in generating not only the primary humoral (B) but also the cellular (T) immune response to the SARS-CoV-2 [22, 26, 27]. These data suggest that the N protein may be a suitable candidate for the development of a vaccine against SARS-CoV-2.
The aim of the research was to obtain the recombinant receptor-binding domain (RBD) of the Omicron variant of SARS-CoV-2 and the nucleocapsid (N) protein of SARS-CoV-2, as well as assess the immunogenicity of the combination of these recombinant proteins in mice. The yeast Pichia pastoris was chosen as a producer of recombinant proteins. Pichia pastoris is a widely used and highly efficient expression system for producing recombinant proteins, making it a popular choice for industrial-scale production. Pichia pastoris is a type of methylotrophic yeast that has several advantages as an expression host for the production of recombinant proteins, including antigens for vaccines [28, 29].
2. MATERIALS AND METHODS
2.1. Bacterial Strain, Yeast Strain, Transfer Vectors, and Materials
The Escherichia coli Top-10F’ (Thermo Fisher Scientific, USA) was used for cloning. The bacterial strain was grown and cultivated in Luria-Bertani (LB) medium (Himedia, India). The LB medium was prepared with 1.5% agar, and the culture was incubated at a temperature of 37°C. The EasySelect™ Pichia Expression Kit containing X33 Pichia pastoris strain; pPICZα A, B, and C transfer vectors; and YPD broth and Yeast Nitrogen Base (YNB) was purchased from Thermo Fisher Scientific (USA). The cultivation of X33 Pichia pastoris was conducted according to the user manual [30, 31]. The restriction endonucleases, T4 DNA ligase, Taq polymerase, DNA ladder, and molecular weight protein markers were obtained from New England Biolabs, Thermo Fisher Scientific, and Sigma-Aldrich (USA). Anti-SARS-CoV RBD (Omicron) and anti-SARS-CoV N (nucleocapsid) antibodies, horse-radish peroxidase (HRP)-goat, and anti-rabbit immunoglobulin G (IgG) antibodies were obtained from Sino Biological (China).
2.2. Cloning of the RBD (Omicron) and N (nucleocapsid) Genes of SARS-CoV-2
The RBD gene of the Omicron variant of SARS-CoV-2, with a size of 707 bp and GenBank accession number MN908947.3, was codon-optimized and synthesized by GenScript Biotech (Singapore) in the plasmid DNA pPICZαA. The gene encoding the N (nucleocapsid) protein of SARS-CoV-2 was amplified from the viral RNA obtained from positive clinical isolates by PCR using synthetic primers. The isolated RNA was subjected to reverse transcription using synthesized specific primers:
F - ATCGGGTACCATGTCTGATAATGGACCCC AAAATCAGCG
R - TGATGCGGCCGCTGCCTGAGTTGAGTCAG CACTGC
The viral RNA sample (5 µL) was mixed with the RT-PCR reagents and the corresponding N gene primers (1 µL, 10 pmol). PCR was performed at 50 °C for 30 min, 95 °C for 5 min, 95 °C for 30 s, 63 °C for 35 s, and 72 °C for 90 s, for 10 cycles. The second step for creating KpnI and NotI restriction sites was performed at 95 °C for 30 s, 69 °C for 35 s, and 72 °C for 90 s, for 30 cycles. Subsequently, the PCR-amplified N gene was identified by employing gel electrophoresis [32].
Primers were synthesized by using the phosphoramidite method and an ASM-2000 synthesizer (Biosset). Plasmid DNA pPICZαA was isolated from E. coli cells by alkaline lysis [32]. 1μg of the pPICZαA transfer vector and the PCR-amplified N gene were sequentially treated with FastDigest KpnI and FastDigest NotI restrictases (Thermo Scientific, USA). After electrophoresis in a 0.8% agarose gel, the treated linear DNA vector with a length of 3536 bp and the N gene was isolated using the PureLink Quick Gel extraction kit (Invitrogen) [33]. Following the ligation of the vector and insert, the ligation mixture was used to transform E. coli competent cells of the plasmid-free Top10F strain (One Shot TOP10F Electrocompetent E. coli, Thermo Fisher Scientific, USA). Agarose gel electrophoresis was performed to determine the molecular weight of the isolated recombinant DNA plasmids obtained from the clones. For a comparative analysis between the recombinant clones and the original vector plasmid pPICZα A, both samples were subjected to restriction digestion at the EcoRI site. Subsequently, the digested DNA fragments were analyzed by electrophoresis on a 0.8% agarose gel to determine the size of the DNA fragments, and a 1 Kb Plus DNA Ladder (Invitrogen) was used as a molecular weight marker.
2.3. Sequencing of Cloned cDNA Fragments
To sequence the recombinant cDNAs, the SeqStudio EDDU protocols for DNA sequencing were followed [34]. The sequencing reactions were performed using the BigDye Terminator v3.1 Cycle sequencing kit (Thermo Fisher Scientific, USA). For each cDNA sample, three identical cycle sequencing reactions were set up. Each reaction contained 30 ng of the template DNA. Since the length of the N gene exceeded 1 kb, it was split into two fragments, and a total of four pairs of primers were used. Each reaction included 10 pmol of gene-specific primers, 4 µL of BigDye Terminator v3.1 ready reaction mix, and deionized water to achieve a final volume of 10 µL. The sequencing reactions underwent an initial denaturation step at 96 °C for 2 minutes. This was followed by 30 cycles consisting of denaturation at 96 °C for 20 seconds, annealing at 55 °C for 10 seconds, and elongation at 60 °C for 4 minutes. The QuantStudio PCR System (Applied Biosystems, Thermo Fisher Scientific, USA) was used for these temperature cycles. Purification of the sequencing products was performed using n-butanol, following the protocol described earlier [35]. Finally, the automated sequencing traces were obtained using the SeqStudio Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, USA).
2.4. Transformation of Recombinant Vectors into P. pastoris
The recombinant vectors pPICZαA-RBD-Omicron and pPICZαA-N were linearized with the SalI restriction enzyme and transformed via electroporation into P. pastoris strain X33. The transformants were plated on yeast extract peptone dextrose medium [YPDS+Zeocin - 1% yeast extract, 2% peptone, 2% dextrose (glucose), 1M sorbitol, 2% agar, 100 μg/ml Zeocin™]. The plates were incubated at 30 °C for approximately 3-5 days until single colonies were observed. The integration of the expression cassette into the yeast genome was confirmed by PCR.
2.5. Fermentation of SARS-CoV-2-RBD Omicron and SARS-CoV-2 -N Recombinant Proteins in P. pastoris
The cultivation of SARS-CoV-2-RBD Omicron and SARS-CoV-2-N recombinant strains of P. pastoris was performed in 30 L Fermenter (Shanghai, China). The initial inoculum, 5–10% of the initial culture volume, was typically prepared in baffled shake flasks containing minimal glycerol (BMGY). BMGY medium contains 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4·10–5% biotin, and 1% glycerol. A culture was initiated with P. pastoris cells from the cell bank and incubated in an orbital shaker at 30 °C, 200-220 rpm, for 16-24 hours until OD600 of 2–6 has been reached. The shake flask culture was transferred to the fermenter, containing 15 L of fermenter medium (BS medium) consisting of the following ingredients per 1 L: H3PO4 85%, 26.7 mL; CaSO4·2H2O, 0.93 g; K2SO4, 18.2 g; MgSO4·7H2O, 14.9 g; KOH, 4.13 g; glycerol, 40 g; and trace element solution (PTM1), 4.3 mL. The PTM1 stock solution contained (for 1 L) CuSO4·5 H2O, 6 g; NaI, 0.8 g; MnSO4·H2O, 3 g; Na2MoO4·2H2O, 0.2 g; H3BO3, 0.2 g; CaSO4·2H2O, 0.5 g L–1; ZnCl2, 20 g L–1; FeSO4·H2O, 65 g L–1; biotin, 0.2 g L–1; and conc. H2SO4, 5 mL. The trace element solution was cold sterilized before use, whereas the BS medium was heat sterilized in the fermenter.
The fermentation process was carried out using the following conditions: the pH was controlled between 5-6 with 25% NH4OH and the temperature was set at 29 ° C. DO was controlled by DO cascade controller between 30% of minimum and maximum rpm. Induction phase was carried out with 1.0% methanol (100% methanol with 12 ml/l PTM1 salts). The methanol induction phase was continued for 72 h [29].
2.6. High-pressure Cell Disruption
Cell disruption was performed using a high-pressure homogenizer (APV Systems SPX 2000 bar Laboratory Homogenizer, USA). Cells were obtained from culture by centrifugation (4,500 x g, 10 min, 4 ºC), followed by two washes of distilled H2O. The cell culture was disrupted for 3 passes at 1200 bar.
2.7. Purification of Recombinant RBD-Omicron and N Proteins
A two-phase separation method was used for the purification of recombinant RBD-Omicron and N proteins from yeast P. pastoris GS115. The recombinant protein containing yeast culture media was recovered by centrifugation at 4500 g for 30 min, and then supernatants were collected for the purification of recombinant proteins. The ammonia sulfate salt (final concentration was 70% for recombinant RBD-Omicron protein and 60% for N protein) was added to the supernatant for isolation of target proteins fraction from unnecessary proteins by centrifugation. Next, fractions containing the target recombinant proteins were dialyzed in special dialysis sacks (Merck, Germany) at +4 °C for 12 hours in the presence of 50 mM sodium phosphate buffer (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole; pH 8.0) using a magnetic stirrer. The recombinant proteins were expressed with a 6×His-tag at the C-terminus to enable purification through Ni2+-charged column chromatography. Subsequently, the desalted fractions of the expressed 6×His-tagged protein were purified using an affinity chromatography column packed with 5 cm2 Ni–NTA resin (Thermo Fisher Scientific, USA). The column was pre-equilibrated with a buffer containing 300 mM NaCl, 50 mM NaH2PO4, and 10 mM imidazole at pH 8.0. After protein adsorption, the resin was washed with 10 volumes of the binding buffer containing 60 mM imidazole (300 mM NaCl, 50 mM NaH2PO4, 60 mM imidazole; pH 8.0). Five volumes of the binding buffer containing 250 mM imidazole (300 mM NaCl, 50 mM NaH2PO4, 250 mM imidazole; pH 8.0) were used to elute the recombinant proteins. All purification steps were performed at 4 °C. The collected fractions were concentrated in a vacuum concentrator (SpeedVac SPD140 DDA) at a temperature of 30 °C and a vacuum of 15 mTorr for 2 h.
2.8. Analytical Assays for Recombinant Protein Characterization
The recombinant proteins were analyzed by SDS-PAGE on a 10% polyacrylamide gel that was stained with Coomassie Brilliant Blue-R250, as described previously [36]. The purified protein concentrations were determined by absorbance at A280 nm and the Lowry method [37].
2.9. Determination of Recombinant Proteins Expression in P. Pastoris
The expression of recombinant RBD (Omicron) and N protein subunits in P. pastoris yeast cells was determined by the standard sandwich enzyme-linked immunosorbent assay (ELISA) method. The SARS-CoV-2 N (nucleocapsid) antibody (Cat. no. 40143-R004, SinoBiological) and SARS-CoV-2 spike antibody (Cat. no. 40150 - D004, SinoBiological) were diluted to 1:5000 with coating buffer (carbonate buffer, pH 9.6) and used as a capture antibody for recombinant N protein and RBD protein, respectively. For this, the diluted antibody was coated onto a flat-bottom 96-well plate (NUNC-MaxiSorp, Thermo Scientific, USA) in 100 μL/well and then left at 4°C overnight. Then, PBS containing 1% BSA (blocking solution) was added to the plate, which was then incubated for 1h at 37°C. After incubation, plates were washed with PBS containing 0.05% Tween 20 (PBST). Then, serially diluted recombinant protein samples were added. The SARS-CoV-2 (2019-nCoV) recombinant nucleocapsid protein (Cat. no. 40588 -V08B) and the SARS-CoV-2 (2019-nCoV) recombinant spike S1+S2 protein (Cat. no.40589-V08B1) were used as positive controls in different dilutions. The plates were incubated at 37 °C for 1 h and then washed five times with PBST. The SARS-CoV/SARS-CoV-2 nucleocapsid antibody (HRP) (Cat: 40143-R040-H, SinoBiological) and SARS-CoV/SARS-CoV-2 spike antibody (HRP) (Cat. no. 40150-D001-H, SinoBiological) were diluted to 1:5000 with PBS containing 1% BSA and used as a secondary antibody. Then, plates were incubated at 37 °C for 1 h and washed 5 times with PBST. The color was developed by the addition of 3.3', 5.5'-tetramethylbenzidine (TMB). The reaction was stopped with 50 μL/well of stop solution (2.0 M H2SO4). The optical density (OD) was measured in a microplate reader (ELx808 Ultra Microplate reader, Biotek Instruments, Inc.) at 450 nm (A450). The cut-off value was defined as A450=0.200.
Moreover, the described method was used for the determination of the recombinant proteins during the purification processes of the proteins.
2.10. Immunization of Animals
The study was carried out on white mice weighing 20-22 grams and, before the experiment, they were kept in quarantine for 1 week in defined standard living conditions. All experimental procedures were approved by the rules of the “European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes” and the Institutional ethical committee in compliance with the Republic Uzbekistan legislation.
The experimental animals were randomly divided into four groups and each group contained 10 animals. The first group was immunized with recombinant N protein (20 μg/150 μl Fruend’s adjuvant), the second group was immunized with recombinant RBD protein (20 μg /150 μl Fruend’s adjuvant) of Omicron variant, the third group was immunized with a mixture of recombinant N protein and recombinant RBD protein of Omicron strain (20 μg +20 μg = 40 μg/150 μl Fruend’s adjuvant), and the fourth group was immunized with 0.9% NaCl/Fruend’s adjuvant (150 μl/150 μl) as a negative control. The animals were immunized three times. In the first and the third injections, complete Fruend’s adjuvant (Thermo Scientific, USA), and in the second injection, incomplete Fruend’s adjuvant (Thermo Scientific, USA) were used as an adjuvant in equal volume (150 μl) with recombinant proteins. Mice were immunized with 300 μl of adjuvant (150 μl recombinant proteins in 150 μl 0.9% NaCl/150 μl adjuvant) to their hind leg muscles on days 0, 14, and 21.
Blood samples were obtained at 25 days post-first immunization by venipuncture from the facial vein. After coagulation at 37 °C temperature for 1h, blood samples were spun in a centrifuge at 3000 rpm/min for 10 min at 4 °C. The upper serum layer was collected and used for ELISA assay of the antibodies against the immunized proteins.
2.11. Analysis of Omicron-RBD and N Protein-specific Serum Antibodies
To detect antibodies against the immunized proteins in the immunized mouse serum, the recombinant Omicron-RBD and N proteins were coated onto a flat-bottom 96-well plate (NUNC-MaxiSorp, Thermo Scientific, USA) at a final concentration of 2 μg/mL (100 μL/well) in coating buffer (Carbonate buffer, pH 9.6), and then left at 4 °C overnight. The levels of antibodies in the third group of animals were determined in two different ways; Omicron-RBD and N proteins were coated onto a flat-bottom 96-well plate individually and together in equal concentration. Then, PBS containing 1% BSA (blocking solution) was added to the plate, which was then incubated for 1 h at 37 °C. After incubation, plates were washed with PBS containing 0.05% Tween 20 (PBST). Then, serially diluted mouse serum was added. The plate was incubated at 37 °C for 1 h and then washed five times with PBST. The goat anti-mouse IgG HRP conjugate (Thermo Scientific, USA) was diluted to 1:80000 with PBS containing 1% BSA and added to the wells. After incubation for 1 h at 37 °C, plates were washed 5 times with PBST and developed with 3.3', 5.5 '- tetramethylbenzidine (TMB) for 15 min. The reaction was stopped with 50 μL/well of stop solution. The optical density (OD) was measured in a microplate reader (ELx808 ultra-microplate reader, BioTek instruments, Inc.) at 450 nm (A450). The cut-off value was defined as A450 = 0.200.
3. RESULTS
3.1. Cloning of the Recombinant Proteins
The transfer vector used for cloning the target recombinant DNA was pPICZα A, having a length of 3593 bp and containing nucleotide sequences from the P. pastoris genome, including the AOX1 promoter. Furthermore, pPICZα contains alpha factor signal sequences (249 bp) that facilitate the secretion of the expressed protein into the nutrient medium [30]. The target cDNA, which encodes the amino acid sequence of the RBD-Omicron or N protein, was introduced into a multi-cloning site. This polylinker consists of various restriction sites, allowing for the insertion of the target genes. In this case, the inserted genes were placed under the control of the AOX1 (alcohol oxidehydrogenase) gene promoter (Fig. 1) [30, 31].
The recombinant subunit nucleocapsid (N) protein, encoded by SARS-CoV-2 nucleotides 28274 to 29530 bp (GenBank: NC_045512.2), was utilized to clone a new recombinant vector plasmid pPICZα A nCoV 2019-N gene (Fig. 1). To achieve this, isolated SARS-CoV-2 genome RNA obtained from positive clinical specimens was employed as the template. The gel electrophoresis results of the amplified cDNA are presented below (Fig. 2).
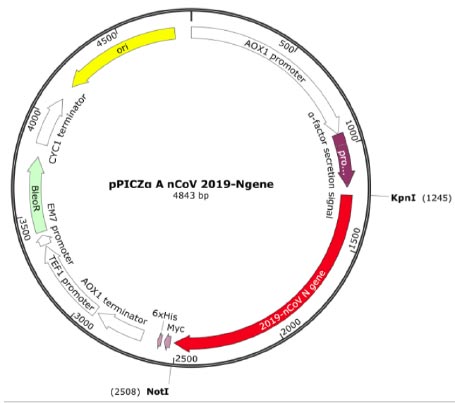
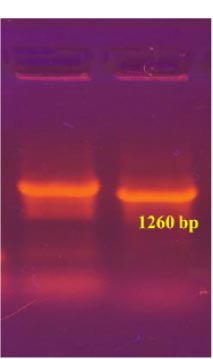
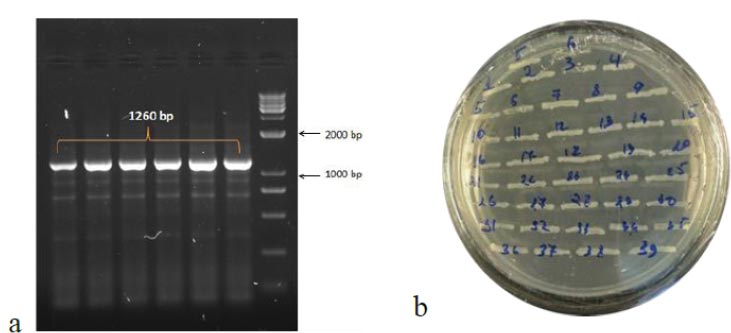
To obtain the recombinant plasmid containing the nucleocapsid (N) gene, the pPICZα A transfer vector and the amplicon from SARS-CoV-2 genome RNA were subjected to sequential treatment with the KpnI and NotI restriction enzymes to create target gene insertion sites in pPICZα A. Following the ligation step, the resulting construct was used to transform E. coli Top10F cells. The recombinant plasmid DNA harbors a gene conferring resistance to Zeocin, enabling the selection of clones (Fig. 3a). The obtained clones were screened to identify those containing the nucleocapsid (N) fragment using PCR with specific primers. The expected size of the PCR amplicon was approximately 1260 bp (Fig. 3b).
In the results of the PCR, DNA fragments of the expected molecular weight indicated the presence of inserts, namely the nucleocapsid SARS-CoV-2 cDNA, within the studied recombinant plasmid (Fig. 3b). Subsequently, in order to obtain P. pastoris yeast strains capable of producing the target recombinant proteins, a transformation (subcloning) process was performed. This involved introducing the recombinant plasmids containing the target Omicron-RBD and nucleocapsid (N) SARS-CoV-2 genes into the X33 P. pastoris yeast strain. To confirm the integration of the target DNA fragments into the P. pastoris genome and to select the recombinant clones synthesizing the target proteins, PCR was employed (Figs. 4 and 5). For identification of the subclone RBD gene, AOX1 P. pastoris genome primers were used, which are as follows:
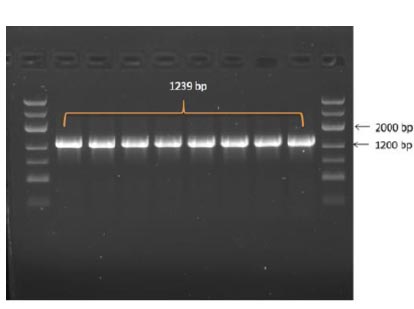
5' Pichia primer - GACTGGTTCCAATTGACAAGC
3' Pichia primer – GCAAATGGCATTCTGACATCC
The anticipated size of the amplicon was 1239 bp (Fig. 4).
The following primers were used to screen the resulting yeast clones containing the nucleocapsid (N) fragment:
F Pichia primer - TACTATTGCCAGCATTGCTGC
R Pichia primer - TGTAGCACGATTGCAGCATTG
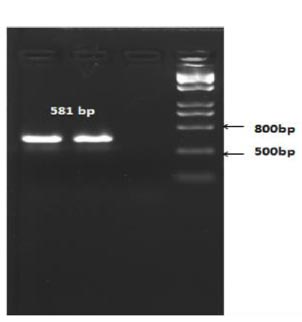
The length of the amplicon was expected to be 581 bp (Fig. 5).
Based on the PCR results, it was determined that recombinant X33 Pichia pastoris yeast strains expressing the RBD-Omicron surface protein and nucleocapsid (N) protein of the SARS-CoV-2 virus were successfully selected.
3.2. Expression of Recombinant Proteins
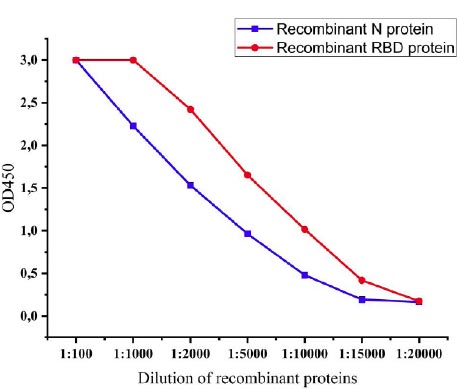
The expression levels of recombinant proteins produced by recombinant P. pastoris strains were determined by the ELISA method. After induction with methanol, levels of expression of recombinant proteins were observed every 6 hours, and at 72 h, the levels of expression reached the highest level. The expression levels of the recombinant N protein and recombinant RBD protein of SARS-CoV-2 were 1:15000 and 1:20000, respectively (Fig. 6).
3.3. Purification of Recombinant Proteins
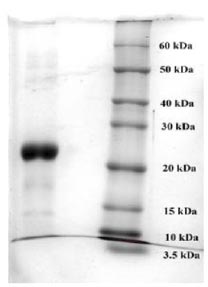
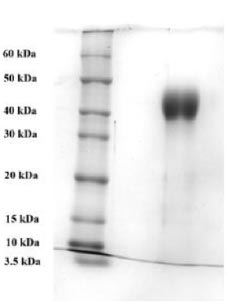
To purify the target recombinant proteins, the yeast cells were homogenized, and the target proteins were precipitated using ammonium sulfate gradient precipitation. The APV-2000 Lab Homogenizer (Germany) was utilized to isolate the target proteins from the yeast cells. The yeast cell culture was passed through the homogenizer under various conditions. The ELISA results indicated that the most optimal condition for the homogenization process was achieved with a 3-fold transmission of samples at a pressure of 1200 bar. In the subsequent step, ammonium sulfate fractionation was employed to purify the target proteins. To remove high molecular weight proteins, (NH4)2SO4 at concentrations between 10% and 25% was used. The ELISA results indicated that salting out samples with 15% ammonium sulfate provided the most optimal condition for the removal of unwanted (ballast) proteins, while minimizing the loss of the target protein. Subsequently, ammonium sulfate solutions at concentrations ranging from 50% to 80% were utilized, and the content of the target proteins in the fractions was assessed using ELISA. Based on the research findings, the optimal salt concentration for the RBD-Omicron protein was determined to be 60%, while it was 70% for the N protein. Following the selection of optimal conditions for the purification of recombinant RBD-Omicron and N proteins through affinity chromatography, we proceeded with purifying the target proteins using the described optimal conditions. Subsequently, electrophoretic analysis of the purified recombinant RBD-Omicron and N proteins was performed using 10% SDS-PAGE (Figs. 7 and 8). SDS-PAGE analysis revealed that the purified recombinant RBD-Omicron and N proteins exhibited approximate molecular masses of 25 kDa and 47 kDa, respectively, which corresponded well with the predicted molecular masses based on the nucleotide sequences.
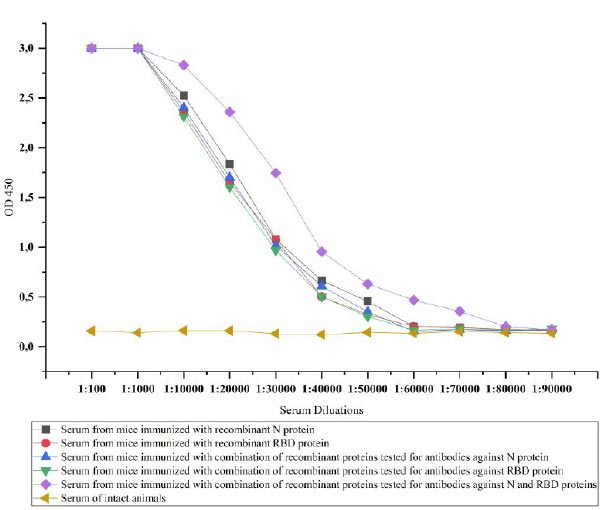
3.4. The Immunogenicity of Recombinant Proteins
The recombinant proteins after purification were injected into the mice to test the ability of immunogenicity. For this, recombinant N and RBD proteins-specific IgG antibodies in the serum of the mice were investigated. IgG is the most common antibody in the fluids of the body and plays a key role in fighting against viral infections [38]. The IgG antibodies specific to recombinant N and RBD proteins in the serum of immunized mice were determined by ELISA. Animal groups immunized with recombinant N and RBD showed similar immunogenicity against injected recombinant proteins until a dilution rate of 1:60000. The immunogenicity was relatively high, i.e., 1:80000 in mice immunized with a combination of proteins, when we simultaneously detected antibodies against the proteins. However, when we tested the serum of animals immunized with the combination of proteins against each protein separately, it was observed that the same level of 1:60000 antibodies formed for both proteins (Fig. 9).
4. DISCUSSION
In this study, the strains of P. pastoris producing recombinant RBD and N (nucleocapsid) proteins of SARS-CoV-2 were obtained. The results of sequencing of the N and RBD genes showed them to correspond to the Omicron SARS-CoV-2 strain. The levels of expression of recombinant proteins in P. pastoris were determined by ELISA. The electrophorese analyses of the purified recombinant proteins were conducted and results showed that the size of the obtained proteins was appropriate to the size of the N (nucleocapsid) and Omicron-RBD proteins of SARS-CoV-2 [39, 40].
The purified recombinant proteins were immunized in the mice to study the immunogenicity of the obtained proteins. Recombinant N and Omicron-RBD proteins and their combination were immunized in mice. The immunization of these two proteins differs from the work of many previous authors because the common vaccines developed against the SARS-CoV-2 virus are based only on the RBD protein [41]. However, it has become clear that the production of vaccines based on only the spike protein (RBD) is not sufficient for defending against the virus. This is due to unprecedented mutations in the S protein of the SARS-CoV-2 virus. In such a situation, it is reasonable to develop new vaccines based on relatively stable or low-mutated proteins of this virus [42]. During the protein immunization process, the animals were divided into 4 groups as follows: animals in the first group were immunized with N protein, animals in the second group were immunized with RBD protein, animals in the third group were immunized with the combination of N and RBD proteins, and the fourth group was the control group. According to the obtained results, the first and the second group of animals showed similar immunogenicity against injected recombinant proteins with a dilution rate of antibody in 1:60000, and the third group of animals exhibited a higher level of antibodies with a dilution rate of 1:80000. In addition, the immunogenicity of the proteins did not decrease even when both proteins were administered together. This means that both proteins can be immunized at the same time as vaccines against SARS-CoV-2. In the studies conducted by previous authors, the presence of antibodies at different levels of dilution can be seen. However, it depends on the titer of antibodies and the ELISA test kits and reagents used to detect antibodies [43].
CONCLUSION
The recombinant RBD-Omicron and nucleocapsid (N) proteins expressed in P. pastoris cells and their combination showed an antigenic specificity and induced the formation of antibodies with strong immunogenicity against SARS-CoV-2 in laboratory animals (mice). The obtained recombinant proteins can be used as the basis for creating new effective vaccines and/or diagnostic tools against new strains of SARS-CoV-2.
Obviously, future vaccines against rapidly mutating viruses, such as Coronaviridae, should be based on two components: the first component should contain surface proteins of the virus containing antigenic determinants, and the second should consist of the most conservative part of the viral genome.
LIST OF ABBREVIATIONS
| SARS-CoV-2 | = Severe acute respiratory syndrome coronavirus 2 |
| RBD | = Receptor-binding domain |
| N | = Nucleocapsid |
| ELISA | = Enzyme-linked immunosorbent assay |
| PCR | = Polymerase chain reaction |
ETHICAL STATEMENT
All experimental procedures were approved by the Institutional Ethical Committee, i.e., Yunusov Institute of the Chemistry of Plant Substances, Molecular Genetics, Tashkent, in compliance with the Republic Uzbekistan legislation. All procedures performed in studies were in accordance with the guidelines “European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes”.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This study was funded by The Ministry of Innovative Development of the Republic of Uzbekistan (grant number: M-2021-5).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
Declared none.


