All published articles of this journal are available on ScienceDirect.
The Hepatitis C Virus NS5A Stimulates NS5B During In Vitro RNA Synthesis in a Template Specific Manner
Abstract
The hepatitis C virus (HCV) NS5B protein contains the RNA dependent RNA polymerase (RdRp) activity that catalyzes the synthesis of the viral genome with other host and viral factors. NS5A is an HCV-encoded protein previously shown to localize to the replisome and be necessary for viral replication. However, its role in replication has not been defined. Using an in vitro biochemical assay, we detected a stimulatory effect of NS5A on the NS5B replication reaction with minimal natural templates. NS5A stimulates replication by NS5B on two templates derived from the 3’ end of the RNA genome (4 fold ± 1.3 fold). A pre-incubation step with the two proteins prior to the replication reaction and substoichiometric levels of NS5A are required for detecting stimulation. With a template derived from the 3’end complementary to the RNA genome (the negative strand) no stimulation was observed. Furthermore, with a synthetic template that allows studying different phases of replication, NS5A stimulates NS5B during elongation. These findings suggest that NS5A stimulates NS5B during synthesis of the complementary (i.e., negative) strand of the RNA genome.
INTRODUCTION
The HCV is a major human pathogen that belongs to the Flaviviridae family of viruses [1]. Understanding how the virus infects cells, replicates its genome, packages and exits the host is critical to developing new strategies to control viral infection. The HCV genome is a (+) strand RNA molecule of ~9.6 kb containing an open reading frame (ORF) and flanking elements at the 3’ and 5’ ends of the untranslated regions (UTRs) [2, 3]. The ORF is translated into a single polypeptide chain that is cleaved by host and viral proteases into ten viral proteins (Structural: C, E1, E2, p7; and non structural: NS2, NS3, NS4A, NS4B, NS5A and NS5B) [3, 4].
NS5B is the RdRP that replicates the viral genome in vitro [5] and in vivo [6]. In vivo replication takes place in a membranous structure derived from the ER in a complex including NS5B, NS4A, NS4B, NS3, and NS5A [7-10] and host factors [7, 11-13]. The carboxy-terminal 21 amino-acid residues of NS5B are important for localization and are predicted to form a transmembrane α-helix [14]. Detailed in vitro NS5B biochemical studies including the processes of nucleotide incorporation, initiation at the 3’ end, abortive and productive initiation, early elongation, stable elongation or termination, have been hampered by the poor ability of the protein to form catalytically competent complexes with the template and NTPs [15, 16]. Contrary to transcription of DNA, replication by HCV does not seem to require a promoter region upstream of the initiation site [17]. With short natural templates from the genomic RNA, NS5B binds to the most terminal region of the 3’UTR [18, 19], and with such templates initiation appears to be the rate limiting step [20]. Additionally, NS5B does not discriminate in vitro between natural and artificial templates, suggesting that additional viral or cellular factors may be required for template specificity. In support of this hypothesis, other viral RNA binding proteins (the helicase NS3 and NS5A) have been shown to be needed for in vivo replication [21].
NS5A is a phosphoprotein essential in the viral infection, including replication [22]. The protein has an N-terminal amphipathic α-helix that serves as a membrane anchor [23] and 3 additional distinct domains [24]. Two isoforms have been detected: one basally phosphorylated and the other hyperphosphorylated [25]. The functional role of different phosphorylation states is not clear [26]; however, loss of hyperphosphorylation in NS5A is reported to stimulate replication [27, 28]. NS5A has RNA binding activity [29], can bind NS5B directly and has been shown to slightly stimulate NS5B catalytic activity with an poly (A) template/poly (U) primer [30]. However, the role of NS5A in replication is still unknown.
In this study, an in vitro replication assay was established to study the role of NS5A. When NS5B and NS5A are pre-incubated, with NS5A at sub-stoichiometric levels, NS5B activity is stimulated up to 5 fold with templates derived from the 3’ end of the (+) strand of viral RNA. No stimulation is detected when NS5B uses a template derived from the 3’ end of the (-) strand of viral RNA. Furthermore, with a synthetic template that allows discrimination between initiation and elongation, NS5A stimulated NS5B during elongation. These results suggest that NS5A acts perhaps by inducing a conformational change in NS5B yielding a more active polymerase with templates derived from the 3’UTR at the elongation phase.
MATERIALS AND METHODS
Materials
Reagents of the highest molecular biology purity were purchased from Sigma Aldrich or Fisher Scientific. Radiolabelled [32P] nucleotides were purchased from Perkin Elmer. NTPs of the highest purity were from GE Healthcare.
Methods
Plasmids Constructs for Template Synthesis or Protein Purification
The plasmid containing the sequence of HCV-Con1 genotype 1b (kind gift of Charles Rice, The Rockefeller University) was used as a PCR template to amplify the sequences of NS5A and all the natural templates. The sequence encoding the HCV full length NS5A (FL-NS5A) was cloned first into the expression vector pSPORT (Invitrogen) cleaved with PstI and XmaI to introduce a 10 histidine tag onto the C-terminus. The primers used to amplify the NS5A sequence were: 10HIS-F: 5’-AAACCTGCAGTCCGGCTCGTGGCTAAG-3’ and 10HIS-R: 5'-GTTGACCCGGGGGCAGCAGACGACGTCC -3'. The NS5A-10HIS sequence was further cloned into the pRSET A plasmid (kind gift of Donald Rio, University of California-Berkeley) digested with AseI and HindIII. The primers used to amplify the DNA from the pSPORT vector were: NS5A-forward: 5’TAACATTCACATATGTCCGGCTCGTGGCTAAG-3’ and NS5A-reverse: 5’ GCGTACGTAAGCTTTTAG-3’. All constructs were sequenced for accuracy.
To generate the templates for RNA synthesis, pMV1 (kind gift of Nam Vo, University of Pittsburgh), which contains the T7 promoter sequence up and downstream of a multiple cloning site, was cleaved with HindIII and NcoI or KpnI. To generate the X (+) template, primers 98XF: 5’-ATCAGCATCAAGCTTAATACAACTCACTATAGGT-GGCTCCATCTTAGCCCTAGTCACGGC-3’ and 98XR: 5’-ATGCGAGTCCATGGACATGATCTGCAGAGAGGCCAGTATCAGCAC-3’ were used to amplify the HCV DNA that was then cleaved with Hind III and NcoI and ligated with pMV1 digested with the same enzymes. To generate the 3’UTR SL (+) template, the forward primer was: 3’UTR-SL F: 5’-ATCAGCATCAAGCTTAATACAACTCACTATAGGGGATCCCCGGCTGCGTCCCAG-3’. The reverse primer was 98XR. Restriction enzyme digestions and ligation with pMV1 were as for the X (+) template.
To generate the 3’ END (-) template (first 341-nt of the complementary strand), the following primers were used: 5’ UTR F: 5’ATCAGCATCAAGCTTAATACAACTCACTATAGGGGATCCGCCAGCCCCCCGATTGGGGGCG-3’ and 5’ UTR R: 5’-CTCCAGAGCATCTGGCACGTGGATGGCTGAGCCATGGGG-3’. The PCR products and pMV1 were cleaved with HindIII and KpnI, and ligated together. All constructs were sequenced after amplification in TOPO10 cells.
The LTMOA template was generated by PCR amplification that combined a T7 RNA polymerase promoter, pBluescript-SK (+) sequence (674 – 914-nt), and a 21-nt G less sequence. The full length sequence of the LTMOA DNA is in supplemental material.
RNA Template Synthesis by T7 RNA Polymerase
The DNA templates for the T7 in vitro transcription reactions were generated from plasmids digested with MlyI [X(+) and 3’UTR SL (+)] or BamHI [3’END (-)]. After purification by gel electrophoresis, phenol extraction and ethanol precipitation, the DNA was dissolved in RNase free water. In vitro transcription reactions were carried out following the supplier directions at 37°C for 4 hours (Epicentre Biotechnologies). After treatment with 0.1U/µL DNaseI for 30 min at 37°C, the RNA was purified by acidic phenol extraction followed by two ethanol precipitations and centrifugation through a Bio-Spin 30 column (BioRad).
NS5A Expression and Purification
Plasmid pRSET-HCV- NS5A-C10HIS was transformed into E. coli BL21(DE3) cells that were plated onto LB medium supplemented with 0.1% Dextrose and 100 µg/mL ampicillin. A single colony was picked after 16-18 hours and inoculated into liquid media for overnight growth. Five mL of the overnight culture were used to inoculate 1 L media. Cells were grown at 37°C with shaking; when the culture reached an OD595nm = 0.8, cells were induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 4 hours at 25°C with vigorous shaking (250 RPM) after first allowing the culture to cool down for 10 min. Cells were then pelleted at 3000×G for 10 min in a SS34 rotor, resuspended in 100 mL ice cold distilled-deionized water and centrifuged again. The pellet was suspended in 25 mL lysis buffer [(Buffer A: 100 mM Tris pH 8.0, 200 mM KCl) with complete-EDTA free protease inhibitors (Roche), leupeptin and pepstatin to a final concentration of 7-10 µg/mL, 2% TritonX-100, and 100 µg/mL lysozyme]. The suspension was quick frozen and stored at -80°C. The FL-NS5A purification was carried out following the Δ32-NS5A protocol [29] with minor differences, including detergent (0.5% Triton X-100) in all buffers.
Pooled fractions containing the peak of NS5A were loaded onto a Superdex - 200 gel filtration column (GE Healthcare) equilibrated in buffer C (50 mM Hepes pH 7.3, 5mM MgCl2, 400 mM KCl, 1mM DTT, 10% glycerol and 0.1% Triton X 100). Two main peaks were observed. Fractions from each peak were flash frozen before storage at -80°C after adding glycerol to 25% final concentration. Protein concentration was determined by the Bradford protein assay (Pierce).
A clone of NS5A lacking the first 32 amino acids (Δ32-NS5A) was obtained from Craig Cameron, University of Pennsylvania, and the protein was purified as described [29].
NS5B Purification
The FL-NS5B was purified as described [31] with minor modifications to increase solubility. Induction with 1mM IPTG was carried out in media containing 2% ethanol and with shaking at 16°C [32] for 16-18H. After Ni-NTA purification, the protein was diluted to 50 mM KCl final concentration with buffer D (50 mM Hepes pH 7.3, 5mM MgCl2, 1mM DTT, 10% glycerol and 0.1% Triton X 100) and affinity purified by using a TSK-Heparin-5PW column (Tooso-Hass) preequilibrated in buffer C with 50 mM KCl. Elution was carried out with a linear salt gradient from 50-700 mM KCl at 0.2-0.5 mL/min. Pooled fractions containing NS5B were concentrated with conical concentrators (5000 MWCO, Vivaspin), flash frozen and stored at -80°C. Protein concentration was determined by the Bradford protein assay (Pierce).
NS5B and NS5A-NS5B Replication Assay
NS5A was diluted in buffer E (50 mM Hepes pH 7.3, 50 mM KCl, 5mM MgCl2, 10µM ZnCl, 1mM DTT, 2% glycerol and 0.1% triton X 100) and then pre-incubated with NS5B for 30 min on ice. NTPs (2mM ATP, 5mM GTP, 1µM UTP, 10 µM CTP), 0.5-1 µM RNA template, 5 U of RNase inhibitor (Invitrogen), and tracer α-32P UTP (5-10 µCi – 3000mCi/mmole, Perkin Elmer) were added in a 10 or 20 µL reaction volume. The replication reaction was incubated at 30°C for 2 hours (unless otherwise stated). To stop the reaction, EDTA (30 mM) and proteinase K (5-10U) were added, and the reactions were incubated at 55°C for 30 min. Then an equal volume of formamide loading buffer (89 mM Trisborate, 0.025% xylene cyanol and bromophenol blue, 18 mM EDTA, 95 % freshly deionized formamide, 0.025% SDS) was added; samples were heated at 95°C for 3 min and loaded onto a 6% (38:2 acrylamide:bisacrylamide) polyacrylamide gel (PAG) containing 7M urea and TBE buffer (50mM Tris-borate at pH 8, 1 mM EDTA). Small gels (16 x 22cm) were run at 20W and large gels (33 x 42 cm), at 80-90W. Gels were exposed to a phosphorimager screen and scanned with a Typhoon 9400 (GE Healthcare). Quantification was achieved by comparison of the product RNA signal to a standard of known concentration of the radiolabeled nucleotide.
Pulse and Chase NS5B-NS5A Reactions
On the LTMOA template, a stalled complex was formed by incubating NS5B or the pre-incubated NS5B+NS5A with a subset of nucleotides (2mM ATP, 5mM GTP and 0.15 µM α-32P UTP) for 30 min at 30°C. Heparin (1mg/mL) was added, and the reactions were incubated for 5 min on ice. CTP and UTP were added to 1 mM, and the reactions were incubated for various times at 30°C. To stop the reaction, EDTA and proteinase K (as above) were added and incubated at 55°C for 30 min. Glycogen (10 µg), 0.3M sodium acetate and 0.1mM EDTA in 100 µL volume were added followed by acidic phenol extraction and ethanol precipitation. The pellet was resuspended into a freshly deionized glyoxal solution: 1.76 M glyoxal, 5% DMSO, in BES buffer (25mM BES, 10mM EDTA, 2mM sodium acetate, pH 6.7). The solution was incubated at 65°C for 50-60 min. Glyoxal loading buffer (BES buffer, 0.025% xylene cyanol and bromophenol blue, 5-7% glycerol) was added, and samples were loaded onto a pre-warmed 10 or 12% PAG containing 7M urea in BES buffer and run at 80-90 W in BES buffer. Gels were fixed (10% acetic acid, 20% methanol) and exposed to a phosphorimager screen as above.
RESULTS
The NS5A protein is essential for HCV replication though its function in the process is unclear [10, 33, 34]. We were interested in testing the effect of NS5A on natural viral sequences. If there were an effect on NS5B, we also were interested in learning about the mechanism.
To test if NS5A had an effect on the NS5B mediated replication, the recombinant bacterially expressed FL-NS5A (Fig. 1A) or Δ32-NS5A (Fig. 1B) that lacks the membrane spanning domain, was used with FL-NS5B (Fig. 1C) during in vitro replication reactions.
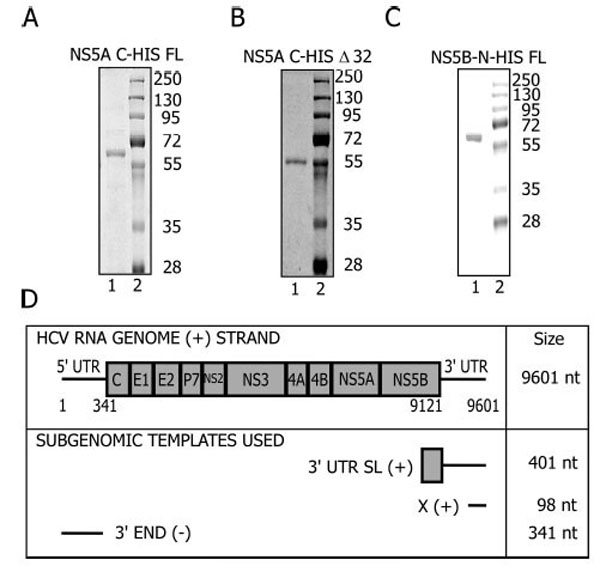
Protein Purification and RNA template design. A. FL-NS5A; B. Δ32- NS5A; C. FL-NS5B Proteins were purified as described in Materials and Methods and resolved on a 10% SDS-protein gel. D. The HCV genome arrangement and the regions used to generate the templates for in vitro reactions.
The templates in these reactions were subgenomic viral RNAs (Fig. 1D) and part of the structural elements of the HCV genome essential for replication in vivo [35-39]. The 3’UTR-SL (+) is a 401 nt RNA template that corresponds to the 3’UTR (+) and sequences in the coding region of NS5B extending from 9212 to 9601. This region contains a stem loop (5BSL3.2) that forms a kissing loop interaction with stem loop 2 of the X (+) element [40] within the 3’UTR. This interaction is essential for in vivo replication [40]. The 3’UTR (+) contains a variable region, a poly (U/UC) tract and the X element [41, 42]. The X (+) template corresponds to the last 98 nucleotides of the 3’ UTR (+) [36], is conserved among different HCV genotypes, is strictly required for viral replication, and folds into three stem loops [36]. The 3’ END (-) template is a 341-nt RNA complementary to the sequence of the 5’UTR of the genomic RNA [43]. NS5B uses this sequence to initiate synthesis of the (+) strand genomic RNA during replication. This template is more efficient in vitro with NS5B than templates derived from the 3’ UTR of genomic RNA [44, 45].
NS5B has low activity during in vitro reactions [5, 46], suggesting a requirement for additional factors to maximize replication efficiency. In initial work with NS5A and NS5B we observed a minor stimulation of RNA synthesis (data not shown) similar to what was observed by Shirota et al. [30]. This stimulation was too small to study.
Thus, we set out to optimize the stimulatory activity starting with the X (+) template. Two conditions proved key. One was pre-incubation of NS5A with NS5B (Fig. 2A, B). When NS5A is added to the reaction with no pre-incubation step, (Fig. 2B lane 2 and Fig. 2C) there is a 1.7 ± 0.4 fold stimulation over the reaction in the absence of NS5A (Fig. 2B, compare lane 1-2 and Fig. 2C). Further addition of the NTP cocktail or the template to the NS5A pre-incubation step did not alter the yield of the reaction (Fig. 2B lanes 3-4 and Fig. 2C). However, when NS5B is pre-incubated with NS5A prior to template addition, a significant stimulation is observed (3.5 ± 1.3 fold; Fig. 2B lane 5 and Fig. 2C). The addition of the template (Fig. 2B lane 6 and Fig. 2C) or the NTP cocktail (Fig. 2B lane 7 and Fig. 2C) to the preincubation of NS5A-NS5B did not significantly alter the stimulation. In summary, NS5A stimulates NS5B replication when the two proteins are pre-incubated prior to the addition of either template or the NTP cocktail.
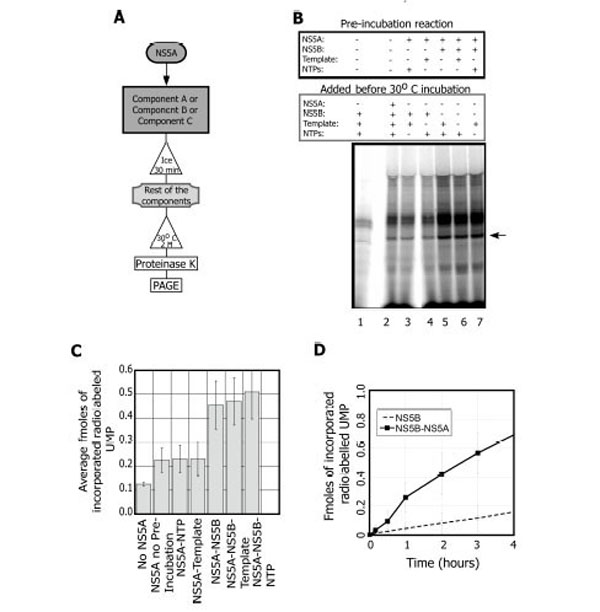
NS5A activates NS5B during the replication reaction in an order of addition dependent manner. A. Scheme of the experimental set up. B. NS5B replication reactions showing components that were pre-incubated on ice for 30 minutes and the components that were added prior to the 30°C incubation. Lane 1, the NS5B (0.3 µM) reaction in the absence of NS5A. Lane 2, NS5A (1 pM) with no pre-incubation prior the replication reaction. Lanes 3-7, replication reactions with NS5A and various pre-incubation additions: NTPs (lane 3); the X(+) template, 0.6 µM (lane 4); NS5B (lane 5); NS5B and the X(+) template (lane 6); and NS5B with NTPs (lane 7). Arrow indicates the expected RNA product. C. Quantification of the effect of NS5A on the replication reaction (average of three independent experiments). D. Time course showing the reactions with NS5B (○) and NS5B+NS5A (■). Each data point is the average of 5 experiments.
The second key condition for maximizing the stimulation of NS5B by NS5A was the use of sub-stoichiometric amounts of NS5A. The stimulation is observed only at very low NS5A concentrations relative to NS5B on this template (0.2 pM NS5A, 0.3 µM NS5B). A reaction timecourse in the presence or absence of NS5A (Fig. 2D) indicated that stimulation continued for at least 4 hours. In summary, when pre-incubated with NS5B, NS5A at sub-stoichiometric concentrations stimulates the replication reaction for up to 4 hours.
Both, the FL- and Δ32-NS5A show similar stimulatory activities. However, the FL is less stable under the conditions used. As a result, we used the Δ32-NS5A throughout the rest of the study. Samples from an identical purification from bacterial extracts induced with a plasmid lacking NS5A had no stimulatory activity, indicating that NS5A is indeed responsible for the stimulation even at such low concentrations. In addition, preliminary mutational analysis supports that the observed effect on NS5B is indeed due to NS5A. Furthermore, in the absence of NS5B no radiolabeled RNA products are detected, indicating that the stimulation is dependent on the NS5A-NS5B interaction, and that NS5A does not have terminal transferase activity or other nucleotide polymerization activity (data not shown). The stimulation conditions were further optimized in 50 mM KCl and 10 µM ZnCl2.
We next evaluated the template dependence of this stimulation. When the 3’UTR-SL (+) template is used (Fig. 3A), similarly to the X (+) template, NS5A stimulates NS5B in a concentration dependent manner (Fig. 3B and C). When NS5A was titrated from 10 nM to 0.1 pM in the replication reaction (0.3 µM NS5B), we again observed that sub-stoichiometric levels of NS5A are optimal for the stimulation (Fig. 3B). The greatest stimulation with this template is observed at 10 pM (4 ±0.6 fold) but 10 times less or more NS5A does not affect the yield greatly (3.8±0.6 or 3.7±1.0 fold stimulation respectively) (Fig. 3B lanes 5-7). Concentrations of 10 nM or higher begin to inhibit the reaction; thus as the amount of NS5A approaches that of NS5B, overall replication in vitro is reduced.
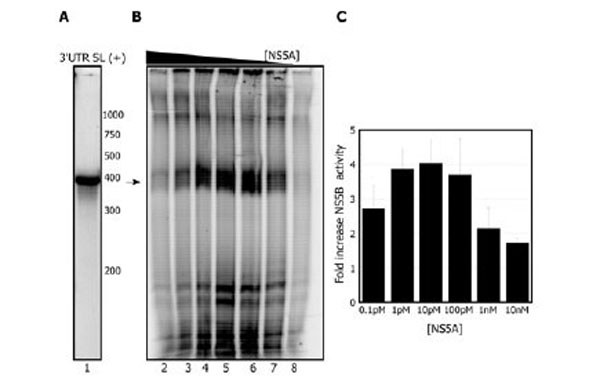
NS5A activates the NS5B replication reaction with the 3’ UTR SL (+). A. Radiolabeled 3’UTR SL (+)template. B. NS5A titration during preincubation with NS5B (0.3 µM) prior to starting the replication reaction: 10 nM NS5A, lane 2; 1 nM NS5A, lane 3; 100 pM NS5A, lane 4; 10 pM NS5A, lane 5; 1 pM NS5A, lane 6; 0.1 pM NS5A, lane 7. Lane 8 shows the reaction with NS5B alone. Arrow indicates the expected RNA product. C. Quantitation of stimulation of NS5B by varying amounts of NS5A. Results are the average of 2 independent experiments.
Templates derived from the 3’ end of the RNA negative strand function more efficiently in vitro with NS5B than templates derived from the 3’ UTR region [44, 45]. With the 3’ END (-) template (Fig. 4A), we also consistently observe 10-100 fold more product with NS5B alone than from templates derived from the 3’ end of the genomic (+) RNA (Fig. 4B). Unlike results with the (+) strand derived templates, we detected no stimulation of NS5B by NS5A with this template. To further test the effect of NS5A, we incubated NS5B at different concentrations with 10 pM NS5A (Fig. 4C) and at varied RNA concentrations (data not shown). In any of these experiments no significant stimulation on NS5B was observed. Taken together these results suggest that the stimulation of NS5B by NS5A is template dependent. These data further suggest that NS5A increases the ability of NS5B to synthesize the (-) strand and not the (+) strand.
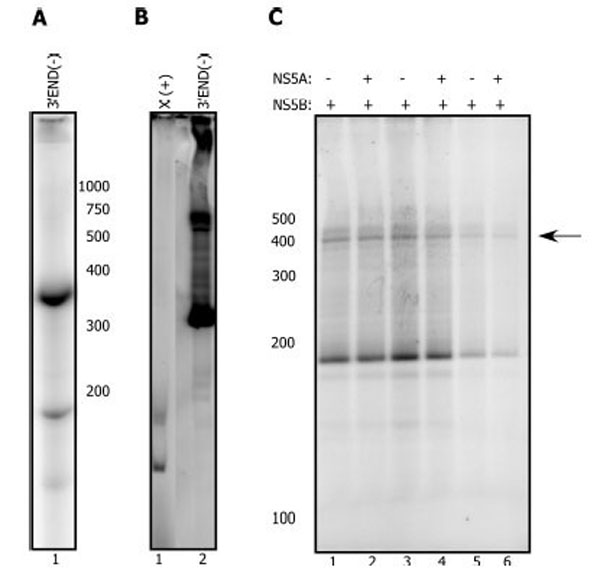
NS5A does not activate the NS5B replication reaction with the 3’ END (-) template. A. Radiolabeled 3’ END (-) template. B. NS5B (2.5 µM) replication reaction with the X(+) template (lane 1) or the 3’ END (-) template (2.5 µM each) C. NS5B (0.3 µM, lane 1, 2) plus or minus pre-incubated NS5A at 10 pM (lane 2). NS5B (0.2 µM, lane 3, 4) plus or minus pre-incubated NS5A at 10 pM (lane 4). NS5B (0.1 µM, lane 5, 6) plus or minus pre-incubated NS5A at 10 pM (lane 6). The reaction proceeded for 2 hours at 30°C after addition of the template (0.2-0.5 µM), and NTPs. Arrow indicates the expected RNA product.
To probe the mechanism of the effect of NS5A, we next tested synthetic templates used previously to study the biochemical properties of NS5B. On one of these templates (LTMOA, kind gift of Dr Nam Vo, University of Pittsburgh), NS5A does stimulate NS5B by 3-5 fold (Fig. 5A, compare lanes 1-2 to 3-4).
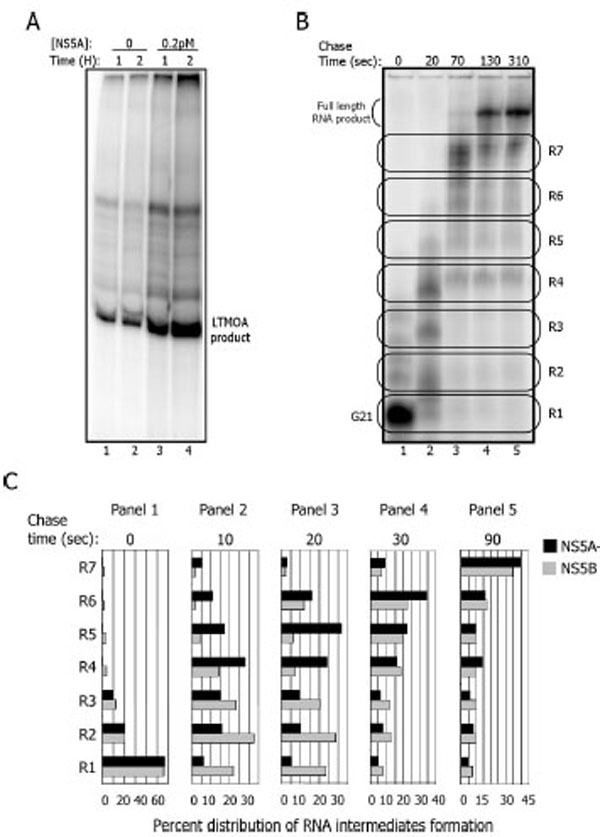
NS5A activation of NS5B occurs during elongation on a synthetic template. A. Replication reaction with the LTMOA template (1 µM) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of NS5A (0.2 pM) for 1 (lanes 1 and 3) and 2 hours (lanes 2 and 4) B. NS5B-containing complexes on LTMOA can be stalled at G21 (lane 1), then released into elongation (lanes 2-5) for 20, 70, 130 and 310 seconds. To quantify the formation of RNA intermediates, the gel was divided into seven regions (R1-R7) for each time point. C. Quantitation of an experiment as in 5B with timepoints of 10 (panel 2), 20 (panel 3), 30 (panel 4), and 90 (panel 5) seconds with and without NS5A.
The LTMOA template allows one to discriminate between RNA binding, initiation, stability of halted ternary complexes, and the elongation phase of RNA synthesis. The sequence of the 3’end of LTMOA does not contain any guanidine nucleotides up to position 21. Thus, in the absence of CTP, NS5B initiates replication and synthesizes RNA up to position 21. Heparin, that prevents re-initiation, and CTP are added to allow elongation to resume. The stalled complex at position 21 (G21) continues synthesis and RNA products can be monitored under single cycle conditions.
To find out how NS5A stimulates NS5B, either NS5B alone or pre-incubated with NS5A is combined with ATP, UTP, GTP, and LTMOA to form G21 complexes (Fig. 5B lane 1). Heparin and CTP are added, and the stalled complexes are incubated for up to 310 additional seconds (Fig. 5B lanes 2-5). To resolve the sizes of the RNA products on denaturing gels, the reactions were treated with glyoxal to prevent secondary structures in the RNA products or template annealing to the products. For data analysis, the RNA products are grouped into 7 regions based upon gel mobility (Fig. 5B). R1 corresponds to the G21 stalled complex, while R2 to R7 identify the accumulation of higher molecular weight RNA products prior to the formation of the full length RNA across a 90 second timecourse (Fig. 5C). The percent of the total products of the replication reaction in each region of the gel in the presence and absence of NS5A indicates that there is no stimulation of NS5B activity in the formation of the G21 complexes (Fig. 5C, R1 panel 1). Rather in the presence of NS5A, the G21 complex elongates its RNA faster into higher molecular weight RNA products than complexes formed only in the presence of NS5B. For example, after 10 seconds, 28% of the RNA molecules are localized at R4 when the reaction is carried out with NS5A, and only 15% of the RNAs are at R4 in the absence of NS5A. After 20 seconds the same pattern is observed (panel 3); 31% of the RNA products are localized in R5 when NS5A is present, while 6% of them are in R5 in the absence of NS5A. No RNase has been detected in these protein fractions, so degradation cannot explain these differential results. In summary these results suggest that NS5A stimulates NS5B during elongation with the LTMOA template.
DISCUSSION
To study the role of NS5A in HCV replication, an in vitro system was set up with recombinant NS5A and NS5B. We found that NS5A stimulates FL-NS5B with either the FL- or Δ32-NS5A. Additionally, detection of the stimulation requires a pre-incubation step and is only achieved when NS5A is present at sub-stoichiometric levels compared to NS5B, as further discussed below.
Co-immunoprecipitation, pull down analysis after fractionation, and immunofluoresence have shown previously that, in vivo, a fraction of NS5A is in a complex with NS5B, and NS3 [3, 47-50]. Previous in vitro studies suggested that (a) NS5A stimulates (20%) the synthesis of a poly U molecule with a 1:10 or 1:50 ratio of NS5A and NS5B, (b) binds directly to NS5B [30] and (c) has RNA binding properties [29]. Yet the specific role of NS5A in replication has remained unknown.
Templates with natural viral sequences were tested and a three to five fold stimulation was observed with the templates derived from the 3’UTR (+). Interestingly, no stimulation was detected with the 3’END (-) template. In addition, with a synthetic template that allows discrimination among steps in replication, we found that NS5A stimulates NS5B activity during elongation.
Significance of NS5B:NS5A Preincubation
The requirement of a pre-incubation step argues that the stimulation is dependent on protein-protein interactions. The NS5A-NS5B protein–protein interaction would not be the first example of a viral RdRP regulated by another viral factor. Birnaviral RdRP (VP1) bound to a peptide fragment of the regulatory viral protein VP3 has been examined for structure and activity [51]. When the peptide fragment of VP3 is pre-incubated with the polymerase (VP1), 3-5 fold stimulation of activity is reported. Furthermore, the activation of VP1 depends upon a conformational change, upon peptide binding, that facilitates the accommodation of the template and incoming nucleotides for catalysis.
Similarly to VP1, NS5B also may require another factor to induce a conformational change. For HCV, the interaction of NS5B-NS5A could lead to the formation of a more efficient NS5A-NS5B complex or alter free NS5B to a more efficient conformation. Perhaps NS5A changes the conformation of NS5B so that the polymerase binds more selectively and replicates more efficiently the 3’UTR (+) derived templates leading to stimulation. This hypothesis is ultimately testable. Indeed, NS5B structural information suggests that the purified enzyme has a “closed” form [52]. A unique feature of the HCV polymerase is a β-hairpin in the thumb sub-domain, which protrudes toward the active site located at the base of the palm subdomain [53, 54]. This hairpin in the closed conformation would be predicted to hinder elongation during synthesis if a template contained a double stranded 3’ end. Indeed, this β-hairpin in the closed conformation blocks the catalytic site when a partially double stranded template is used [55]. When part of the β-hairpin is deleted, NS5B is able to initiate the synthesis of RNA on this template [55, 56]. It is possible that NS5A induces movement of the NS5B β-hairpin or another regulatory domain allowing the accommodation and elongation of templates. NS5B, once in its open conformation, might recognize the template without any additional aid from the factor, and this conformation may also stimulate elongation.
Template Selective NS5A Stimulation of NS5B
In vitro, purified NS5B has a very low rate of nucleotide incorporation (2-4 nt/sec) [46] with templates derived from the 3’ UTR (+) as well as with synthetic RNAs. This elongation rate contrasts with 20-200 nt/sec for a variety of other RNA polymerases [57], including a reported 70 nt/sec for poliovirus RdRP [58]. However, NS5B uses the 3’END (-) template very efficiently in vivo [50] and in vitro ([44, 45] and this work), although the mechanism of this template specificity is not known. NS5A stimulates replication by NS5B on templates derived from the 3’ UTR (+) that have a double stranded 3’ end and on at least one synthetic template with a single stranded 3’ end, yet no stimulation is detected on the 3’ END (-) template that also has a single stranded 3’ end. This result suggests that the template character of the replication start site is not rate limiting in the in vitro stimulation by NS5A. Since NS5A has no effect on 3’ END (-) replication, it is possible that the 3’END (-) template itself enables more efficient elongation either intrinsically due to its sequence or because this template itself promotes a conformational change in NS5B that contributes to more efficient replication.
The results with the natural templates are consistent with the hypothesis that NS5A enables NS5B to recognize essential elements of the RNA genome and stimulates the synthesis by NS5B of the negative strand over the positive strand. Thus, in vivo, one role of NS5A might be the regulation of template utilization by NS5B.
Replication Elongation
Using the LTMOA template, we found that NS5A was able to stimulate NS5B during elongation. NS5A does not decrease the number of pause sites recognized by NS5B, but rather changes the kinetics of RNA intermediate formation. The LTMOA template has a 3’ end predicted to be free of secondary structure so that NS5B might bind and initiate efficiently. Clearly NS5A does not affect the formation of the G21 stalled complex on this template under these conditions. However, as the polymerase resumes elongation, structural or sequence elements in the template might require the influence of NS5A on NS5B for efficient elongation. This hypothesis is supported by the observation that NS5A does not stimulate NS5B catalytic activity on three other synthetic templates even though the first 21 nucleotides are identical to those in LTMOA. Two of these three templates have a 3’ end free of predicted secondary structure (data not shown). This result again suggests that the rate limiting step impacted by NS5A is not initiation.
Stimulation at the elongation phase is observed when NS5A is pre-incubated with NS5B prior to the formation of the G21 complex. Whether stimulation is also achieved when NS5A is depleted from the G21 complex before elongation or when the factor is added after stalled complex formation is still under investigation. These experiments will reveal whether NS5A forms a complex with NS5B to stimulate its catalytic activity or whether a conformational change in NS5B induced by the factor can be achieved once the polymerase is in the ternary complex. The low stoichiometry required for detecting stimulation by NS5A suggests that it might act catalytically on NS5B; future experiments will examine this possibility.
With the analysis presented here, we propose that NS5A binds NS5B and induces a conformational change that “opens” NS5B. The open form might better accommodate elongation on defined templates. Structural studies of the NS5A-NS5B complex and additional biochemical analysis should expose the molecular details of the mechanism of stimulation.
In summary, by devising conditions to study the in vitro effect of factors in the HCV replication reaction, we have identified a role for NS5A in stimulating replication. The mechanism of stimulation might involve the rotation of the NS5B β-hairpin or other regions of NS5B that block the catalytic site creating a conformation that is elongation competent. Further studies are needed to detail this effect of NS5A. Of course, in vivo replication occurs with a complex of host and viral factors in addition to NS5A and NS5B. The conditions presented in this study make it possible to explore the role of additional proteins in replication. These biochemical studies coupled with in vivo studies will enable a clearer understanding of HCV replication and its regulation.
SUPPLEMENTARY MATERIAL
Supplementary material can be viewed at Download Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr. Nam Vo and Salvatore Cilia for their scientific input and critical discussion. EQ would like to thank Dr. Stuart Linn for critical discussion and help with data analysis.
ABBREVIATIONS
REFERENCES
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]


