All published articles of this journal are available on ScienceDirect.
Characterization of the Cdc6 Homologues from the Euryarchaeon Thermoplasma acidophilum
Abstract
Archaeal cell division cycle protein 6 (Cdc6) homologues are thought to be involved in the initiation process of DNA replication. In the present study, a biochemical characterization of the two Cdc6 proteins from the archaeon Thermoplasma acidophilum has been performed. Both TaCdc6-1 and TaCdc6-2 behave as monomers in solution and both are abundantly expressed in vivo. Further, TaCdc6-1 shows strong ability to undergo autophosphorylation compared to TaCdc6-2 and the autophosphorylation activity is not affected by DNA or MCM
INTRODUCTION
In archaea, homologues to eukaryotic Cdc6 proteins are thought to be involved in origin recognition and loading of the replicative DNA helicase prior to DNA replication (reviewed in [1,2]). The Cdc6 proteins are members of the AAA+ (ATPases associated with various cellular activities) family [3] and crystal structures have shown that they have a N-terminal ATP binding module consisting of two domains and a C-terminal winged-helix (WH) domain [4-7]. The Cdc6 proteins and other initiation proteins are called AAA+ switches due to their ability to alter their structure and function depending on the resident nucleotide, and thus, the most important mode of regulating the DNA replication is through nucleotide binding and hydrolysis (reviewed in [2,8]).
All archaeal Cdc6 proteins studied to date have shown the ability to undergo autophosphorylation and it has been suggested that autophosphorylation might play a regulatory role during the initiation process [9]. In addition to autophosphorylation, archaeal Cdc6 proteins are able to bind both single-stranded (ss) and double-stranded (ds) DNA and some Cdc6 proteins bind specifically to origin of replication (reviewed in [1,2]). Although the archaeal Cdc6 proteins are monomers in solution, co-crystallization of the proteins with inverted DNA repeats found within the archaeal origins of replication and footprinting analysis, have shown that they interact with origin DNA as dimers [4,10].
Thermoplasma acidophilum is a thermoacidophilic euryarchaeon encoding two putative Cdc6 homologues (TaCdc6-1 and -2) and one putative minichromosome maintenance (MCM) homologue, TaMCM. Previous studies have shown that TaCdc6-2, but not TaCdc6-1, interacts with TaMCM and stimulates its helicase activity [11,12]. The work presented here is a study on the TaCdc6 homologues, including size determination, in vivo levels of expression and their ability to autophosphorylate.
MATERIALS AND METHODS
Cloning and Protein Purification
The pET21a - constructs with Ta0451m (encoding TaCdc6-1) and Ta0636 (encoding TaCdc6-2) were made as described previously [11]. The recombinant proteins were over-expressed in Escherichia coli BL21-CodonPlus (DE3) RIL cells (Stratagene) using ZYP-5052 medium according to Studier’s method for auto-induction [13] and purified as described previously [11].
Gel Filtration
For molecular mass determination 72 μg of each of the purified TaCdc6 proteins were applied to a superdex 200 gel-filtration column (HR10/30; GE Healthcare) pre-equilibrated with buffer A containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl and 10% glycerol. The gel filtration experiments were performed at 22°C with a flow rate of 0.5 ml/min.
Sedimentation Velocity Measurement
Sedimentation velocity experiments were done on a Beckman XLI analytical ultracentrifuge, equipped with a UV scanning system, using a four holes AN-60 Ti rotor with double–channel centrepieces of 1.2 cm optical path-length. The protein sample (0.25 mg/ml) was in buffer B containing 50 mM Tris, pH 8.0. Two hundred absorbance profiles at 280 nm for each sample were recorded at 42,000 RPM at 20°C. The experimental sedimentation coefficients were determined using the Sedfit software [14].
Autophosphorylation
Protein autophosphorylation activity was measured in reaction mixtures (15 μl) containing 20 mM Hepes-NaOH, pH 7.5, 5 mM MgCl2, 2 mM DTT, 450 ng BSA, 3.3 pmol of [(-32P]ATP (GE Bioscience), 10 pmol TaCdc6 proteins with or without 1 μg ssDNA ((X174 virion DNA; New England Biolabs) or dsDNA ((X174 RFII DNA; New England Biolabs) and with or without 20 pmol TaMCM as indicated in the figure legends. Following incubation at 58°C for 20 min, the reaction was stopped by adding 5 μl of 5 x SDS loading buffer (250 mM Tris-HCl, pH 6.8, 500 mM DTT, 10% SDS, 0.5% Bromophenol blue and 50% glycerol). The reaction mixtures were boiled for 5 min and subsequently separated by 12% SDS-PAGE performed at 190 V for 60 min and stained with Coomassie blue. The gel was dried and 32P-labeled bands were detected using phosphorimaging. The experiments were repeated three times. A representative gel and autoradiograph is shown in Fig. (4A and B).

A.The activity of TaCdc6 is not affected by DNA. Cdc6 autophosphorylation reactions were performed as described in “Materials and Methods”in a reaction mixture (15 μl) containing 10 pmol Cdc6 protein in the absence (lane 2 and 5) or in the presence of 1 μg ssDNA (lane 3 and 6) or 1 μg dsDNA ((lane 4 and 7). Upper panel shows a representative 12% Coomassie stained SDS-polyacrylamide gel. Lower panel shows a representative autoradiogram.
B.Cdc6 autophosphorylation reactions were performed as described in “Materials and Methods” in a reaction mixture (15 μl) containing 10 pmol Cdc6 protein in the absence (lane 3 and 5) or presence of 20 pmol MCM (lane 4 and 6). Upper panel shows a representative 12% Coomassie stained SDS-polyacrylamide gel. Lower panel shows a representative autoradiogram.
Cultivation of Thermoplasma Acidophilum
T. acidophilum (DSM 1728) were grown in DSMZ medium 158 (http://www.dsmz.de/microorganisms/html/media/ medium000158.html), pH 1.9 at 55°C with shaking and the cells were harvested in exponential phase. The growth was monitored by OD measurements at 600 nm and cell counts as indicated in Fig. (3A).
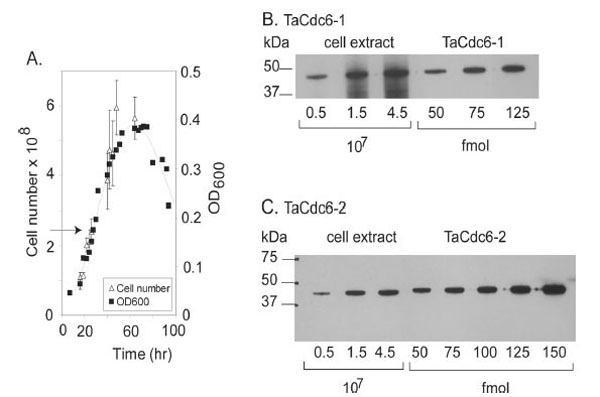
A.Growth curve of T. acidophilum. The number of cells (108) and density (OD600) are plotted versus time (hours). The arrow indicates the time of harvesting.
BQuantitative Western blot of TaCdc6-1 inT. acidophilumwas performed as described in “Materials and Methods”. T. acidophilumextract corresponding to a certain number of cells (x 107) and amount of purified, recombinant TaCdc6-1 protein (fmol) loaded onto each lane are indicated.
C Quantitation of TaCdc6-2 in T. acidophilum cells was performed as described in B.
Quantification of TaCdc6 Proteins Using Western Blot Analysis
Purified, recombinant TaCdc6 proteins and cell extract from T. acidophilum were fractionated using 12% SDS-PAGE, followed by electro-blotting onto a nitrocellulose membrane. Western blot analyses were performed using TaCdc6-1 and TaCdc6-2 polyclonal antibodies generated against the recombinant proteins by BioGenes GmbH (Germany), and developed using enhanced chemiluminescent (ECL, GE Healthcare). The bands were quantified using Image J [15].
Modeling
The models of TaCdc6-1 and TaCdc6-2 were generated by automated homology modeling using Deepview/Swiss-PdbViewer v3.7 [16,17]. The FASTA sequence of TaCdc6-1 was fitted to 1FNN, the crystal structure of Cdc6 from Pyrobaculum aerophilum (PaCdc6) [6], suggested as template using the “magic fit” application of the program, while TaCdc6-2 was fitted to 2QBY which is the structure of Cdc6 from Sulfolobus solfataricus [4]. The resulting preliminary alignment was manually adjusted to align residues corresponding to the active site and other conserved areas based on ClustalW2 [18] alignment before submission to Swiss-Model Automated Comparative Protein Modeling Server (http://swissmodel.expasy.org/) [16,17,19].
RESULTS AND DISCUSSION
The TaCdc6 Proteins are Monomers in Solution
Purified TaCdc6 proteins (Fig. 1) were analyzed by gel filtration for mass determination. TaCdc6-1 eluted at 14.3 ml which corresponds to a monomer having a theoretical molecular mass of 42 kDa (Fig. 2A). TaCdc6-2 was expected to elute at approximately the same volume as it has similar theoretical molecular mass (46.5 kDa). TaCdc6-2 did, however, elute at 13.2 ml, the same volume as albumin which has a molecular mass of 67 kDa (Fig. 2B). A limitation of gel filtration as a method for molecular mass determination is that it does not account for shape and hydration of proteins and thus, the method will work well only if the protein of interest has a spherical symmetrical shape and an average hydration level [20]. The behavior of TaCdc6-2 was thus investigated using analytical ultracentrifugation in order to monitor the oligomeric state and shape of the protein. Under the conditions described in “Materials and Methods”, the theoretical value of a globular monomeric TaCdc6-2 is 3.7 S. The experimental sedimentation profiles were analyzed using the continuous c(s) distribution model with bimodal frictional coefficient (f/fo) in the sedfit program. The data were well fitted by a mixture of globular (50%) and extended (50%) monomeric forms without dimer (data not shown). The globular monomeric form (f/f0 = 1.25) has an experimental s value of 3.6 S, i.e. close to the theoretical value. The second form has a lower s value of 2 S and a frictional coefficient f/f0 = 1.5 showing that the monomer also exists in an extended shape. The equilibrium between the extended and globular forms explains the TaCdc6-2 retardation using the gel filtration column. The results agree with previous studies reporting that archaeal Cdc6 proteins are monomers [6,7,21-23]. However, X-ray diffraction crystallography has shown that the Cdc6-1 and -3 homologues in Sulfolobus solfataricus (SsCdc6-1 and -3) form a hetero-complex at the origin of replication [4]. Whether this is the situation in T. acidophilum and other archaea remains to be determined.
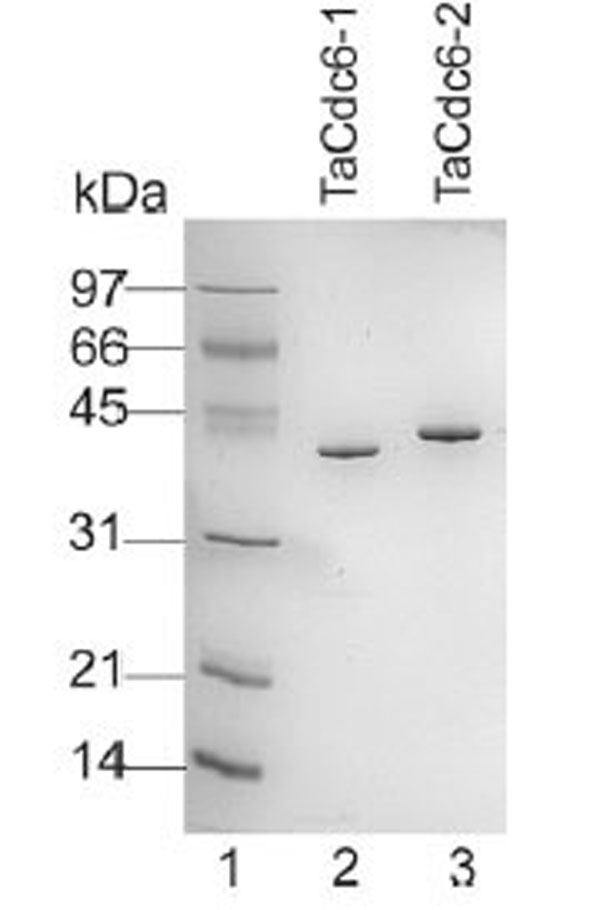
Aliquots (0.5 μg) of each of the purified TaCdc6 proteins were fractionated on a 12% SDS-PAGE gel and visualized by Coomassie blue staining. Lane 1: molecular mass marker; lane 2, T. acidophilum Cdc6-1; lane 3, T. acidophilum Cdc6-2.

A. Gel filtration chromatography of TaCdc6-1 was performed as described in “Materials and Methods”. The peak elution volumes of thyroglobulin (Tyr, 670 kDa, 8.76 ml), ferritin (440 kDa, 10.02 ml), catalase (Cat, 232 kDa, 11.7 ml), albumin (Alb, 67 kDa, 13.22 ml), ovalbumin (Ova, 45 kDa, 14.34 ml) and myoglobulin (Myo, 13 kDa, 16.56) are shown in the elution profile of TaCdc6-1 (upper panel) and used to make a calibration curve (lower panel). The arrow indicates the position of the TaCdc6-1 protein
B. Gel filtration chromatography of TaCdc6-2. The position of molecular marker proteins is shown as described in A. The arrow indicates the position of the TaCdc6-2 protein.
TaCdc6-1 and TaCdc6-2 are Abundantly Expressed in Thermoplasma Cells.
In order to verify the expression of the putative TaCdc6 homologues in vivo, polyclonal antibodies against recombinant TaCdc6-1 and TaCdc6-2 were raised in rabbits. Western blot analyses of a cell extract of T. acidophilum revealed specific bands corresponding to similar molecular mass as the recombinant proteins, indicating that both proteins are indeed expressed in vivo. A quantitative Western analysis was subsequently performed in order to determine the number of the TaCdc6 proteins per cell during exponential growth phase. The intensities of bands corresponding to varying numbers of Thermoplasma cells were compared with different molar amounts of recombinant TaCdc6 proteins (Fig. 3B and C), suggesting the presence of 3000 - 4000 molecules per cell of each of the two TaCdc6 proteins. This is in agreement with observations from other archaea [21,24]. The reason for the large number of Cdc6 proteins in archaea is currently unknown, but it is suggested that the Cdc6 proteins may have other functions in the cell in addition to regulation of DNA replication. For example, studies of S. solfataricus have shown that the transcription of SsCdc6-2, but not SsCdc6-1 and -3, was highly induced in UV treated cells, implicating a function in DNA repair [25,26].
The TaCdc6 Homologues are Able to Undergo Autophosphorylation
Both TaCdc6-1 and TaCdc6-2 contain conserved AAA+ motifs as well as winged-helix (WH) motifs as shown by sequence comparison analysis. Also, models of TaCdc6-1 and -2 three-dimensional structures were constructed, and as expected, both were similar to other Cdc6 proteins (data not shown). As both proteins belong to the AAA+ family, ATPase activity was expected. However, only weak ATPase activity was observed (data not shown). This is similar to other archaeal and eukaryotic Cdc6 proteins [9,21,27].
The ATPase activity of Cdc6 proteins can, however, also be indirectly shown by their ability to autophosphorylate. This is because the proteins have to bind ATP and break the interaction between the (β and (γphosphate prior to phosphorylation [9]. To investigate whether the TaCdc6 proteins are able to autophosphorylate, the proteins were incubated at 58°C as described in “Materials and Methods” in the presence of ((γ32P(ATP. As shown in Fig. (4A) (lane 2 and 5), TaCdc6-1 undergoes stronger autophosphorylation than TaCdc6-2. Although both proteins show ability to autophosphorylate, the level of autophosphorylation is low. In other archaeal Cdc6 the level of phosphorylation was measured to be less than 1% [9].
Previous studies with Methanothermobacter thermautotrophicus, have shown that DNA affects the Cdc6 autophosphorylation [9]. However, as shown in Fig. (4A) (lane 2 versus lane 3-4 and lane 5 versus 6-7), the activity of the TaCdc6 proteins is not significantly affected by DNA. This is similar to the Cdc6 homologues in S. solfataricus, suggesting that the regulation of autophosphorylation varies within the archaeal domain.
Both archaeal Cdc6 and MCM proteins are suggested to be involved in initiation of DNA replication and studies have shown that the proteins affect each others biological activities (review in [1,2]). Thus, the effect of the TaMCM onTaCdc6 autophosphorylation was analyzed. A TaMCM mutant, K343A, where lysine in the conserved Walker-A motif has been replaced by alanine, is devoid of ATPase activity [11] and used in this experiment as ATP hydrolysis by MCM would limit the available ATP. Otherwise, the TaMCM wild-type and K343A mutant are similar and both interact with TaCdc6-2 [11,12]. As shown in Fig. (4B) (lane 3 versus 4 and lane 5 versus 6), TaMCM did not significantly affect the TaCdc6 autophosphorylation activity. This is different from studies with M. thermautotrophicus where the Cdc6-MCM interaction modulates the autophosphorylation activity of the Cdc6 proteins [28].
All archaeal Cdc6 proteins studied have shown the ability to autophosphorylate. The in vivo function of autophosphorylation is not yet known, but it has been suggested that phosphorylation might play a regulatory role during the initiation process. This regulation might either be positive, allowing Cdc6 binding to origins, or negative, preventing re-binding of Cdc6 to the origins once the helicase is loaded into DNA [9,29]. A mechanism of preventing re-replication (re-initiation of a recently activated origin within the same cell cycle) in eukaryotes is to limit the accessibility of initiation proteins by protein phosphorylation, and thereby “labeling” the proteins for degradation or export out of the nucleus. In this context it has been suggested that autophosphorylation might be a mechanism to prevent re-replication in archaea [29].
In addition to possessing different biochemical properties, as reported here and in a previous study [11], TaCdc6-1 and TaCdc6-2 belong to different subgroups of Cdc6 [7,30]. It is thus tempting to speculate whether the two Cdc6 homologues have separate roles during the initiation of DNA replication in T. acidophilum. However, further analysis such as DNA binding is needed to address this question.
REFERENCES
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]


