All published articles of this journal are available on ScienceDirect.
Biochemical and In-silico Studies on Pectin Methylesterase from G9 Variety of Musa acuminata for Delayed Ripening
Abstract
Ripening of fruit is a very important process but in some fruits early ripening leads to a great damage during long distance transportation. There are various biochemical changes taking place during the phase of ripening of fruit such as changes in respiration, aroma, flavor, ethylene production and activity of cell wall degrading enzymes. Some important cell wall degrading enzymes are Polygalacturonase (PG), Pectin methylesterase (PME), Pectin lyase, RGase. PME is known to act as a cell wall hydrolyzing enzyme, responsible for demethyl esterification of cell wall polygalacturonan. The present study includes the biochemical and molecular characterization of PME from Grand naine variety of Musa acuminata (banana). This study also deals with the in-silico study reflecting inhibition of PME activity in context to delayed ripening in banana. It mainly deals with the identification of a PME1 gene from Grand naine variety of banana. The expression of this gene is related with the process of ripening. The expression of PME1 gene was observed to be peaked on 3rd day in ethylene treated samples of banana but the activity in untreated samples called control was rather slow and then there was a sudden decrease in their activity in both treated as well as untreated samples. With the help of in-silico study, we observed that banana has maximum homology with carrot by using cross species analysis.The designed model has been reported to be of good quality on the basis of its verification and validation. The designed model was observed to be appropriate for docking. The information of binding sites of ligand provides new insights into the predictable functioning of relevant protein.
INTRODUCTION
Banana is a very economical and important crop belonging to Musa species. It plays an important role in providing nutrition due to being rich source of carbohydrates and millions of population depends on banana for the source of food [1]. The genome of banana may be diploid or triploid but commonly having AAA type genome such as Cavendish and is parthenocarpic [2]. The banana plant is monocot and ripening of fruit is influenced by ethylene. On fruit ripening, several biochemical changes take place such as change in carbohydrate, cell wall attachment, production of volatile compounds and degradation of chlorophyll [3]. Due to the presence of high content of starch and phenolic compounds, the protein extraction and determination are more difficult in case of banana.The supply of ethylene is necessary for complete maturation as the rate of respiration increases during ripening. The color of peel as well as the taste of pulp becomes changed on ripening of banana. The starch content is converted into sugar and thus the taste becomes sweeter of
ripen fruit. The textural changes, maturation and softening occur due the presence of pectin in the cell wall, which has been reported to decompose into sugar and acids during ripening [4].
The cell wall of plants consists of polysaccharides mainly pectin [5]. The pectin is well known to be composed of distinct group of polymers such as rhamno- galacturonan I, rhamno- galacturonan II and homogalacturonan [6]. The textural properties of middle lamella present in cell wall were reported to base on cross linkages of adjacent pectin molecules and on polymerization and esterification of polygalacturonides [7]. The strength of unripen fruit has been reported due to the presence of protopectin or water insoluble pectin, which probably having been partially esterified polygalacturonic acid [8]. The pectin was noticed to convert into soluble form on ripening and the chain length also decreased and thus the texture becoming softer and softer. The bridge of α- (1→4)-linked D-galacturonic acid constructs the methyl esterified pectin chain. The increase in soluble pectin concentration and softening of fruit is related with the activity of pectic enzymes. Pectin degrading enzymes are PG and PME [9]. Generally, PME is used to remove the methyl ester groups and then depolymerization takes place by polygalacturonase PG [10]. The extraction and purification of PME have been documented from different plants including tomatoes [11], oranges [12, 13], apples [14, 15], and grapefruits [16]. The increased activity of PME has been shown in banana [17], orange [18], tomato [19] and strawberry [20] and it remained constant in banana [21], tomato [22], and mango [23] but decreased in tomatoes [24], avocado [25] and mango [26].
Globally, there are several varieties of banana known for their large economic value. We selected the Grand naine variety of banana for this study. Grand naine is also known as Williams’s hybrid or giant Cavendish due to their high yield. It is a cultivar of Cavendish banana with AAA triploid genome. It is parthenocarpic and produces seedless fruit [27]. It lacks sexual reproduction and fast grower plant and having longer shelf life. The flavor of this variety is supposed to be richer and thus containing a firm fleshy fruit. Itcan bear up to 90 bananas in bunches and can withstand in hardly and windy environment. On ripening, the covering of fruit becomes thick, smooth and yellowish green in color. The taste of fruit is sweet with fine textured and pleasant aroma. Due to the tolerance to abiotic stresses and good quality of fruit, Grand naine variety of banana may soon become the most preferred one.The objective of this hypothesis was to determine the significance of PME in relation to fruit softening and to characterize the expression of PME in Grand naine variety of banana along with their in-silico molecular docking study to identify the inhibitor against PME, to increase the availability of banana fruit.
MATERIALS AND METHODS
Plant Material and Sample Preparation
The mature unripen banana of Grand naine variety was collected from the Biotech Park Lucknow, India. To initiate the ripening in selected banana samples, ethylene treatment was given at a concentration of 100µl in a closed chamber for 24 hours. After ethylene treatment, samples were allowed to ripen at room temperature for 6 days and collected at unripen, mid-ripen and fully ripen stage on 1st, 3rd and 6th day, respectively of both ethylene-treated as well as untreated samples (control). The tissue of banana pulp was crushedunder the condition of liquid nitrogen and kept at -80⁰C till further use.
Firmness of Fruit
The firmness of sampled fruits was monitored on 0-day, 1st, 3rd and 6th day of both treated and control samples. The measurement of firmness was recorded as Newton (N) by Penetrometer (model FT 327, QA Supplies, Norfolk VA).
Pectin Extraction and Estimation
The total content of polysaccharides of plant cell wall was obtained as solids with alcohol insolubility method [28] followed by further isolation of pectin molecules from this fraction as water soluble pectin (WSP), chelator soluble pectin (CSP) and HCl soluble pectin (HSP) [29].The crushed banana pulp samples were mixed with ethanol and boiled for 35 min. On filtration, the residue obtained was washed twice with absolute ethanol. Fifty mg of this residue was dissolved in 60 ml of distilled water and kept overnight incubating at 20⁰C with continuous stirring. The residue obtained after filtration was washed with 5 ml of distilled water for two times and the obtained filtrate was called as WSP. After filtration, the remaining residue was treated with 50 ml of EDTA of 0.05M. The filtrate obtained after filtration called as CSP and after treated the residue with 0.05M HCl at 100⁰C for 60 min, then the filtrate obtained was designated as HSP fraction. In all fractions i.e. WSP, CSP and HSP, the concentration of galacturonic acid (GA) was measured by m-hydroxydiphenyl method [30]. The pectin content was expressed as nmolGAg-1AIS.
Enzyme Extraction and Assay of PME
One g of tissue pulp was crushed with 3 ml of PBS (1X) buffer, prepared homogenate and then centrifuged at 15,000 X g for 30 min at 4°C. The protein content was assayedaccording to Lowry et al. [31]. The activity of PME was calculated volumetrically using the following formula:
PME units/ml = (ml of NaOH) (molarity of NaOH) (1000)
(time) (ml of sample)
The mixture was prepared by 15 ml of 0.25% of pectin solution in 0.15M NaCl. Two hundred µl of sample wasadded, made up final volume of 30 ml with distilled water and pH adjusted to 8.0. The mixture was then incubated at 30⁰C for an hour, followed by treatment with 0.1M NaOH. Phenolphthalein was used as an indicator dye.
RNA Extraction, Reverse Transcription and PCR
Ribonucleic acid was extracted from all the banana samples with the help of CTAB method [32]. After the formation of RNA, complementary DNA or c-DNA was synthesized by reverse transcriptase (Invitrogen, Carlsbad, CA) by using oligo dT primer. To amplify the PME gene fragment from banana, primer PME F1 5’CTTTTACCG-CAGGGTTGA 3’ and 3’AP primers were employed.
Sequence Retrieval and Cross Species Analysis
The sequence of amino acid of PME of banana was retrieved from the entrez protein database [33] followed by specific multiple sequence alignment by using default parameters predicting the homology among all ten plants as described in Table 1 and by the study of phylogenetic tree evaluation given in Fig. (1), it was clear that the banana PME was shown maximum homology with carrot PME.
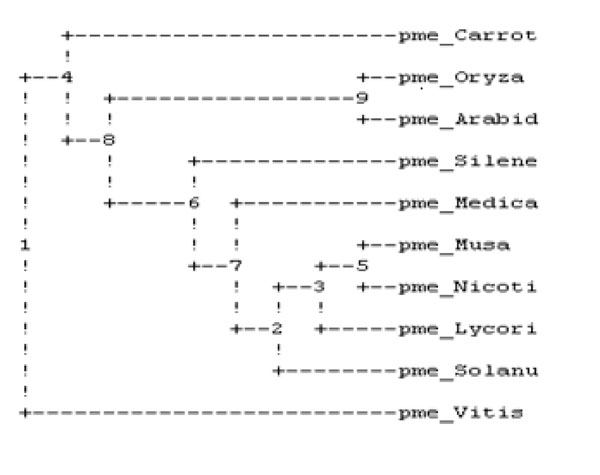
Phylogenetic analysis of ten plant’s PME.
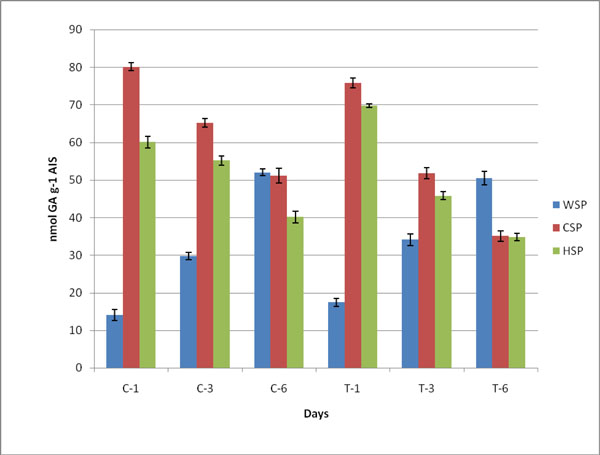
Changes in pectin, WSP, HSP and CSP in both control (C) and ethylene treated (T) samples during ripening of banana fruit for 1st Day (C1 and T1), 3rd Day (C3 and T3) and 6th Day (C6 and T6). Values are mean ± SD of three sets of experiments with triplicates in each set.
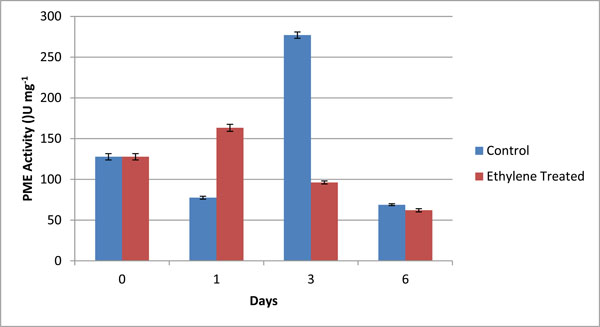
PME Specific activity (U mg-1) in different fractions of cell wall from control (C) and ethylene treated (T) at day 1, day 3 and day 6. The graphical data are mean of 3 sets of experiments. Values are mean ± SD of three sets of experiments with triplicates in each set.

mRNA extracted from both control (C) and ethylene treated (T) on Day0 (control i.e. C0=T0), Day1 (unripe- C1and T1), Day3 (midripe -C3 and T3) and Day6 ( fully ripened -C6 and T6).

Expression of PME after PCR from all control i.e. C0- control, C1-unripe, C3-midripen, C6- fully ripen and ethylene treated, T1-unripe, T3- mid-ripen and T6- ripen fractions of banana with AP marker.
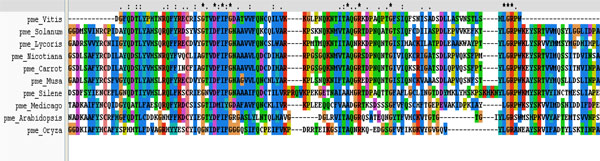
Multiple sequence analysis shows the three conserved motifs present in the PME of different plants.
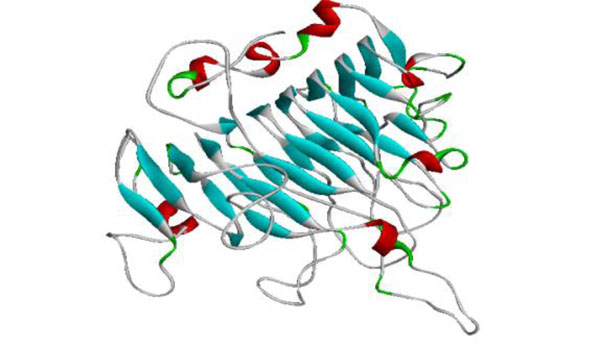
The generated model of PME.
Plants selected for multiple sequence alignment.
| S.No. | Plant name | Accession Number |
|---|---|---|
| 1. | Solanumtuberosum | CBY44654.1 |
| 2. | Nicotianabenthamiana | AAO85706.1 |
| 3. | Mendicagotruncatula | CAB65290.2 |
| 4. | Arabidopsis thaliana | CAB87932.1 |
| 5. | Silenelatifolia | CAA69348.1 |
| 6. | Vitisvinijera | AAD45347.1 |
| 7. | Lycorisamea | ABJ99595.1 |
| 8. | Oryza sativa | AAQ20039.2 |
| 9. | Daucouscarota | 1GQ8_A |
Firmness changes during ripening in banana for 0, 1, 3 and 6 Days, for ‘E’ ethylene treated fruits and ‘C’, control fruit samples.
| DAY | CONTROL | ETHYLENE TREATED |
|---|---|---|
| 0 | 12 ± 2 | 12 ± 2 |
| 1 | 11.23 ± 0.18 | 11.16 ± 0.15 |
| 3 | 10.76 ± 0.26 | 3.16 ± 0.12 |
| 6 | 10.33 ± 0.15 | Nd |
Changes in pectin (nmol GA g-1 AIS) in different fractions of cell wall from controland ethylene treated at day 1, day 3 and day 6. The values are mean ± SD of 3 sets of experiments with triplicates in each set.
| Day 1 | Day 3 | Day 6 | ||||
|---|---|---|---|---|---|---|
| C2H4 | Control | C2H4 | Control | C2H4 | Control | |
| WSP | 17.5 ±1.05 | 14.1 ±1.40 | 34.5 ± 1.49 | 29.8 ± 1.01 | 50.5 ±1.79 | 52.0 ±0.90 |
| CSP | 75.8 ±1.31 | 80.1 ±1.05 | 51.8 ± 1.51 | 65.2 ± 1.12 | 35.1 ±1.40 | 51.2 ±1.91 |
| HSP | 69.8 ±0.55 | 60.1 ±1.61 | 45.8 ± 1.05 | 55.2 ± 1.21 | 34.8 ±1.01 | 40.1 ±1.56 |
| Total Pectin | 165.5 ±1.02 | 150.2±3.15 | 140.6 ± 1.59 | 148.5 ± 1.32 | 120.1 ±1.40 | 149.2 ±4.28 |
PME activity (U/ml) and relevant specific activity (U/mg protein) in different fractions of cell wall from control (C) and ethylene treated (T) at day 1, day 3 and day 6. The values are mean ± SD of 3 sets of experiments with triplicates in each set.
| S.no. | Sample | PME activity (U/ml) | Specific unit (U/mg protein) |
|---|---|---|---|
| 1. | C-0 | 100.00 ± 4.50 | 127.71± 3.91 |
| 2. | C-1 | 73.33± 1.83 | 77.51± 1.73 |
| 3. | C-3 | 106.66± 3.63 | 277.01 ± 3.81 |
| 4. | C-6 | 60.01 ± 1.21 | 68.88± 1.18 |
| 5. | T-1 | 40.05 ± 0.95 | 163.26± 4.27 |
| 6. | T-3 | 40.10 ± 1.01 | 96.15± 1.85 |
| 7. | T-6 | 86.66 ± 2.36 | 62.07 ± 1.97 |
Calculated various energies of the different inhibitors.
| S. No. | Ligand Name | BEe | Vdw-Hb-Ds | Ic | Td | Eb | Calculated Ki |
|---|---|---|---|---|---|---|---|
| 1. | 1Methyl Cyclo Propane | -2.27 | -2.27 | -2.27 | 0.0 | 0.0 | 21.62 mM |
| 2. | Salicylic Acid | -4.69 | -3.59 | -0.35 | +0.82 | -1.57 | 363.50 uM |
| 3. | Gallic Acid | -6.40 | -5.71 | -0.46 | +1.37 | -1.60 | 20.39uM |
| 4. | NAA | -2.26 | -2.26 | +0.00 | +0.00 | +0.00 | 22.08 mM |
| 5. | Vanillic Acid | -2.25 | -2.87 | -0.34 | +1.10 | -0.14 | 22.27mM |
| 6. | PME1 | -1.46 | -1.61 | -0.84 | +1.92 | -0.93 | 84.92mM |
| 7. | Salicin | -9.29 | +4.59 | +1.12 | +2.47 | -9.86 | 0.00 M |
| 8. | Ferulic acid | -1.45 | -2.62 | -0.37 | +1.37 | +0.17 | 87.22mM |
| 9. | Cinnamic acid | -5.23 | -4.41 | -0.04 | +0.82 | -1.59 | 147.78 uM |
| 10. | Green Tea Catechin extract | -9.05 | +0.00 | +3.45 | +3.29 | -9.40 | 0.00 M |
| 11. | Indole -3 Acetic Acid | -5.20 | -5.40 | -0.21 | +0.82 | -0.41 | 155.51uM |
| 12. | Ethanol | -2.21 | -2.33 | +0.00 | +0.27 | -0.15 | M |
Multiple sequence analysis was performed across ten plants by using ClustalW (http://www.ebi.ac.uk/Tools/ msa/clustalw2/) software using default parameters, which depicts the conserved regions of proteins. Accession numbers of all ten plants were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/) as summarized in Table 1.
Selection and Designing of Macromolecule
Daucous carota was employed as a parent template for designing the 3-D structure pertaining to PME of Musa acuminata by using Modeller9v8. The selection of template was accomplished by protein BLAST [34] using PDB database [35]. The best designed model for PME was selected on the basis of DOPE and lowest RMSD (root mean square deviation) by superimposing on template.Energy modifications were accomplished by using Swiss PDB Viewer (spdbv) software [36].
Model Validation and Submission
The quality of generated 3-D structure of PME was ensured by using Structural Analysis and Verification Server (SAVES) [37].The evaluation and validation of generated model were executed with PROCHECK [38], ERRAT [39], WHAT-IF [40], PROSA [41] and VERIFY3D [42]. Chimera [43], PyMolv0.99 [44] and Discovery Studio Visualizer2.5 [45] softwares were used for visualizing the 3D structure of proteins of interest.
Retrieval and preparation of Ligand database
By using Pubchem database [46], twelve inhibitors of ripening were retrieved along with their qualitative and quantitative properties. The file format was converted by using Open Babel software [47].
Molecular Docking
Molecular docking studies were performed with selected compounds by using Lamarckian genetic algorithm inbuilt in Autodock4.2 tools with default parameters.The grid map was generated by using a grid of 46*40*52 spanning the binding pocket of the protein with the grid point spacing of 0.38. The grid map covered the active site with surrounding surface [48]. Grid parameter files were built and atom-specific 3D affinity maps were constructed using Autogrid4. The binding energies were calculated by considering the three steps of docking finally projecting with a population size of 150 yielding 10 docked conformations. Accordingly the best docked conformation was accepted with minimum energy calculations.
Molecular Interaction and Binding Assessment
The binding energies were determined by estimated inhibition constant (Ki), which was calculated by using Autodock 4.2. To estimate the binding energies in kcal mol-1 the electrostatic energy, Vander Waals, hydrogen bond, desolation energy, total intermolecular and torsional energy of binding were used. Chimera, Discovery Studio3.1 and PyMol softwares were used for the visualization of interactions.
RESULTS
Firmness
As a consequence of ethylene treatment the ripening of Grand naine variety of banana was observed to be rapid. The force of penetration decreased from 12N to 3.16N within three days. The untreated samples also showed a decrease in penetration force from 12N to 10.76N. On day six, the penetration force was 0 due to fully ripened fruit Table 2.
Pectin Degradation During Ripening in Banana
Duringthe process of ripening in banana, the pectin degraded and was estimated as WSP, CSP and HSP Table 3. The decrease in content of pectin showed pectin solubilization on ripening. The content of uronic acid increased in WSP and the level of GA significantly increased during ethylene induced banana ripening. The amount of soluble uronide was maximum on 6th day. The contents of CSP and HSP were observed to be decreased (Fig. 2). The value of pectin degradation was higher than control samples.The covalently bound pectin decreased whilewater soluble pectin increased ultimately leading to softening. The contents of chelator and HCl soluble pectin were monitored to decrease on ripening of banana fruit (Table 3; Fig. 2). It was observed that in both cases the content of WSP increased while CSP and HSP decreased due to the solubilization of pectin during ripening of fruit.
PME Activity in Banana
The activity of PME was assayed by volumetric analysis in the pulp of banana during ripening, and calculated in terms of specific units i.e. U/mg. The values obtained by volumetric analysis were shown in Table 4 and Fig. (3). The activity of PME was measured in grand naine variety of banana during ripening. The activity was measured for both control as well as ethylene treated samples on day 1, day 3 and day 6. Day 0 was considered as control for both types of samples. On day1, the activity of PME in ethylene treated samples wasobserved to increase abruptly and in control ones was also increased but at slower rate because exogenous supply of ethylene involved in ripening more rapidly as compared to natural synthesis (Fig. 3). On day 3, the activity of control samples increased more rapidly due to the activity of ethylene formation whereas in treated samples, it decreased and on day 6 there was no significant difference between both control and treated samples (Fig. 3). It was predicted that during ripening of banana, the activity of PME increased initially and then declined.
Expression Exploration of PME in Banana
mRNA was isolated from different stages of ripening i.e. unripen, mid-ripen and fully ripen from all control and treated samples. As shown in Fig. (4), there was a high transcript level in C1, C3, T1 and T3 as compared to C0, C6 and T6. The high quality and expression of RNA was assessed by RT-PCR probed with 24 bp fragment of PME1 gene (Fig. 5). On ripening, the transcript level in ethylene- treated as well as control sample increased reaching up to maximum on day 3 and then decline. Further, the expression of PME gene in grand naine variety of banana was monitored to enhance initially with ripening time and then decline after some time. This proves that PME, a cell wall degrading enzyme, helps in softening of cell wall and promotes the ripening of fruit till a peak time followed by decline. In Fig. (5), C1 band showed more expression than C0, C3 and C6 and similarly the expression level of T3 band was more than T1 and T6 because at mid-stage i.e. C1 and T3 in both control as well as ethylene treated fruit respectively, the activity of PME increased and then declined. The control for both fractions were taken same (T0=C0).
Cross Species Analysis
It was performed on ten plant varieties to assess the conserved regions and to search a template for developing 3D structure of PME1 (Table 1). The result of multiple sequences showed the presence of conserved regions containing motif in various PMEs (Fig. 6). These conserved regions at the active site of protein are involved in the binding of inhibitors. Interestingly, the phylogenetic analysis of PMEs from different plants (Fig. 1) depicts the maximum homology of banana with Daucous carota (Carrot).
Model Generation
The template chosen for model generation was PME of Daucous carota (PDB ID: 1GQ8) on the basis of maximum similarity. Five models were generated and the model showing the least RMSD with respect to trace (Cα atoms) of the crystal structure of the template, optimal DOPE and GA314 score was selected for further validation. The generated model showed the enzyme to have 7 alpha helices and 24 beta sheets. The high quality model of Musa acuminata has been submitted to pmdb database and has pmdb id PM0078247 (Fig. 7).
Interaction of Ligand with Protein by Docking Studies and Energy Calculations
Molecular docking was performed by using Auto Dock 4.2 tool on 12 inhibitors for the prediction of interaction between PME1 protein in banana and each inhibitor. The optimal docking energy conformation of each ligand, their inhibitory constant and binding energies values have been shown in Table 5.
The present study depicts that Asp381 residue is the major interacting residue involving in binding of substrate with enzyme. The lowering of the value of Ki is directly proportional to the docking energy and inversely proportional to the binding affinity.
Where, Eb: electrostatic content of binding free energy in kcal/mol.
Vdw-Hb-Ds: Vander Waals, Hydrogen bond-Desolvation energy component of binding free energy in kcal/mol.
Ic: Total internal energy in kcal/mol.
Td: Torsional energy in kcal/mol.
Ki: Inhibition Constant in molor.
BEe: Estimated binding free energy in kcal/mol.
Molecular docking studies entail that there were various ligands or inhibitors binding with the active site pocket of PME. For the good binding of ligand and proteins, the binding energy of ligand should be low and the value of inhibitory constant zero. The best inhibitor of PME1 gene was selected on the basis of interaction at the active site as well as optimized energy values. Based on energy calculations for protein inhibitor interaction, out of twelve inhibitors, the binding energy of only two were observed to be very low and thus were designated as good inhibitors for gene PME1 in grand naine variety of banana.
Firstly some inhibitors could be suggested as good inhibitors such as Gallic acid, IAA, Salicylic acid and Green tea catechin because they all have low binding energies but few of them did not show any interaction at the active site. Ethanol and 1-MCP were not included because they both were having very high binding energies. Gallic acid, IAA and Salicylic acid did not show any interaction on the active site although having good binding energies.
The protein inhibitor interaction was shown by the complexes formed by Salicylic acid, IAA, Gallic acid, Cinnamic acid and ethanol but at Ser387 which was other than active site, and thus discarded. At Arg470 V motif of LGRPW i.e. one of the active region, the pi-interactions were showed by PMEI, Vanillic acid and Ferullic acid but they were also not considered because of their high energies. Salicin and Green tea catechin showed the interaction at Asp381 of 3rd residue of YQDTL i.e. one of the active site region and both of them having very low binding energies, and thus could be considered as good inhibitors.The binding energy of Salicin and Green tea catechins are very low and are considered them as good inhibitors.
DISCUSSION
The objective of this investigation was to course the expression of PME from Grand naine variety of banana followed by the identification of the best inhibitor of PME through in-silico studies to increase the more availability of banana for poor population to provide carbohydrate rich nutritious food. A large amount of fruits especially perishable fruits such as banana becomes damaged during transportation from one place to another and there is a huge loss of both economy as well as nutrition on which a large population is dependent. The present study reflects that at the time of ripening of fruit the activity of PME increases and then decreases. Other studies also show that on ripening various changes take place but the activity of pectin degrading enzymes (PG and PME) increases and lead to hydrolyze the pectin, ultimately, responsible for softening of fruit. To establish the hypothesis of delaying the ripening of banana fruit, we performed the biochemical, molecular and in-silico studies.
In biochemical studies it was shown that the firmness of ethylene treated samples became ripened at very fast rate as compared to control samples, reflecting ethylene to play an important role in ripening. Also, at ripening, the content of WSP was reported to be increased and CSP as well as HSP to be decreased. The activity of PME increased initially and then abruptly declined in both cases.
Based on the present molecular studies, it was registered that on ripening the transcript level increased and through RT PCR the expression of PME was monitored to be maximum at C1 and T3, because of being at the starting time of ripening and then the expression declined as C6 and T6. With the help of these studies itcan be concluded that if there is delayed activity of PME, the ripening of fruit would also be delayed up to some extent and ultimately the availability of fruit becomes increased. In in-silico studies, by cross species analysis, the conserved regions of different fruit’s PME could be analyzed through Phylogenetic analysis, it was predicted that the banana PME had maximum homology with carrot PME and therefore, carrot was chosen as the template for further modeling and docking studies.The best model was generated on the basis of validation and verification, which was further used for molecular docking with different inhibitors. Various inhibitors were selected and through docking their interactions with target protein PME could successfully be analyzed.The protein ligand interaction reflects that that there are two inhibitors i.e. salicin and Green tea catechin, which can be considered as good inhibitors on the basis of their low binding energies’ calculations as well astheir binding at the active site. Taken together the previous study [49] and the present one, it can be confirmed that at low concentration the activity of salicin is always less effective in delaying the ripening of tomato but at high concentration it is more effective. Green tea catechin or epigallocatechin gallate is a natural inhibitor of PME, which specifically blocks the PME activity [49]. Both salicin and green tea catechin show their interaction at Asp381 of DFIFG 4th motif, which is one of the active regions of PME. Some other inhibitors also have low binding energies but they do not show any interaction with the active site pocket and hence were discarded in the final concluding study. Conclusively, green tea catechin was established as the best inhibitor due to salicin being concentration dependent whereas green tea catechin as a natural inhibitor. Nevertheless, green tea catechin is the best inhibitor to inhibit the activity of PME in banana to delay their ripening. The findings of this study provide new insights into the probability of increasing the availability of banana fruit by inhibiting the activity of PME concomitant with new lines of synthesizing genetically modified banana fruits
CONFLICT OF INTEREST
The author declares no funding for this project, and no conflict of interest.
ACKNOWLEDGEMENTS
The present study is a part of Ph.D. program of author (CV) who is registered at Uttarakhand Technical University, Dehradun, UK, India.


