Unique Photobleaching Phenomena of the Twin-Arginine Translocase Respiratory Enzyme Chaperone DmsD
Abstract
DmsD is a chaperone of the redox enzyme maturation protein family specifically required for biogenesis of DMSO reductase in Escherichia coli. It exists in multiple folding forms, all of which are capable of binding its known substrate, the twin-arginine leader sequence of the DmsA catalytic subunit. It is important for maturation of the reductase and targeting to the cytoplasmic membrane for translocation. Here, we demonstrate that DmsD exhibits an irreversible photobleaching phenomenon upon 280 nm excitation irradiation. The phenomenon is due to quenching of the tryptophan residues in DmsD and is dependent on its folding and conformation. We also show that a tryptophan residue involved in DmsA signal peptide binding (W87) is important for photobleaching of DmsD. Mutation of W87, or binding of the DmsA twin-arginine signal peptide to DmsD in the pocket that includes W72, W80, and W91 significantly affects the degree of photobleaching. This study highlights the advantage of a photobleaching phenomenon to study protein folding and conformation changes within a protein that was once considered unusable in fluorescence spectroscopy.
INTRODUCTION
Reduction of dimethyl sulfoxide (DMSO) to dimethyl sulphide is catalyzed by the redox enzyme DMSO reductase. During anaerobic respiration in bacteria, DMSO serves as a final electron acceptor in the electron transport chain. DMSO reductase is comprised of the DmsABC heterotrimer, where DmsAB is targeted to the DmsC membrane anchor in the cytoplasmic membrane via the twin-arginine translocation (Tat) system [1]. DmsA contains a typical Tat-signature motif in its signal peptide that is important for enzyme assembly and Tat-dependent targeting to the cytoplasmic membrane [2]. The signal peptide is cleaved and removed from the final holoenzyme upon targeting and translocation, presumably by leader peptidase I LepB [3].
Maturation of DMSO reductase requires the help of a system-specific chaperone, DmsD [4], which is encoded on a separate operon from dmsABC,ynfIEFGHI, where ynfI was renamed dmsD [5]. DmsD has been shown to interact with the twin-arginine (RR)-containing DmsA signal peptide [6-10], likely to aid in cofactor insertion, folding, and targeting as these are generalized roles for redox enzyme maturation protein (REMP) chaperones [11-13]. DmsD has also been shown to interact with the TatBC subunits of the translocase [14,15], potentially to transfer DmsA(B) for translocation. It belongs to a family of REMP proteins that include TorD, NarJ, NarW, and YcdY [11,12,16]. Several crystal structures of DmsD and its homologs are available [17-19]. It is mainly α-helical in structure, with an identified surface pocket suggested to be important for binding to the DmsA signal peptide [18-20].
Fluorescence spectroscopy is a useful technique in studying proteins. It can be used to monitor protein folding [21,22] and protein-protein interactions in vitro and in vivo [22,23]. Intrinsic fluorescence of proteins relies on intrinsic fluorophores – the aromatic residues tryptophan, tyrosine, and phenylalanine, but mainly tryptophan as it has the largest quantum yield and Stokes shift properties [24]. Fluorescence quenching can occur when the fluorophore interacts with its environment in a way to have its excited state energy relaxed resulting in decreased fluorescence. Photobleaching occurs when the fluorescence decrease is irreversible due to chemical change of the fluorophore. Photobleaching can be useful in some types of experiments such as fluorescence recovery after photobleaching (FRAP) experiments (reviewed by Ishikawa-Ankerhold et al. [25]). However, it is often frustrating in an experiment when one is interested in utilizing fluorescence spectroscopy to study folding and ligand binding of biomolecules. The fluorescence intensity gradually decreases due to photobleaching during repetitive or continuous exposure to light at the chromophores’ excitation wavelength, leaving the protein inaccessible to many types of studies.
Here we characterize the self-quenching and photobleaching effect in DmsD. We find that DmsD is severely and irreversibly photobleached upon irradiation at the typical excitation wavelengths used to study Tyr and Trp environments in proteins. Although this was frustrating to our planned experiments, we explored this phenomenon to understand why it is so dramatic in this protein. We found that this effect is directly correlated to the folding and conformation of the protein. Identification of Trp residues modified upon irradiation showed that several of these residues are located in the DmsA RR-signal peptide binding pocket on the surface of its structure. We found that binding of DmsA signal peptide by DmsD had significant effects to the photobleaching in DmsD, demonstrating that binding alters its conformation to expose tryptophan residues, especially those suggested to be associated with RR- signal peptide binding (W72, W80, W87, and W91).
MATERIALS AND METHODS
Materials
DmsD wildtype or W87S mutation variant was expressed and purified as described in Winstone et al. [26], and stored at -20 ᴼC. Prior to use, purified aliquots of DmsD were thawed on ice and then vortexed for 10 s and then centrifuged at 10,000 rpm for 30 min. Trititachium album Proteinase K was obtained from Sigma-Aldrich. DmsA signal peptides corresponding to residues 2-20 (DmsA2-20) or 15-41 (DmsA15-41) purchased from Custom Peptide Synthesis services (Genscript) were used as described by Winstone et al [26]. All other reagents are as described by Ladner et al. [27].
Fluorescence Spectroscopy
Fluorescence spectra were collected on protein samples for 10 μM DmsD-Trp in 50 mM ammonium bicarbonate buffer, pH 8.0 at 20 °C, unless otherwise stated. Spectra were collected with a Fluorolog FL-211 spectrofluorimeter (Horiba Jobin-Yvon Spex) equipped with a 450 W Xenon lamp and controlled by DataMax v2.0. Samples of 2 ml were excited at 280 nm and emission was recorded from 300 to 500 nm, with a 1 s integration time. The excitation and emission band passes were 2 nm for all experiments. Temperature was controlled using an RTE-111 (Neslab) water bath circulating through the cuvette holder. All readings were recorded with a 1 cm path length quartz cuvette.
After a first scan to collect the entire spectra, samples were excited at 280 nm for 1000 s and emission was collected at 350 nm at increments of 1 s. At the end of a time course, the entire emission spectra were collected from 300-500 nm. Initial quenching rates over the first 30 s of irradiation were calculated using the following equation:
The rate r is directly proportional to the difference between the initial fluorescence intensity F0 at time t0, and the final intensity F after time t. This rate accounts for the initial velocity of reaction, which shows a linear fluorescence decrease at the beginning of the irradiation. The decrease of intensity within the first 30 s of the experiment was found to be linear. For every reaction R2 of the fit curve was found to be > 0.95.
According to the Stern-Volmer equation, the quenching constant Ksv is correlated to the concentration of the quencher. In our case, no external quencher was present, but a decrease of fluorescence was observed upon continuous irradiation. Thus in our study the variable is represented by the time of exposure. Therefore, we describe the photobleaching in the equation below to provide a constant to compare experiments. The initial fluorescence intensity F0 at time t0 was measured after the sample temperature had equilibrated. The sample was then irradiated with steady light exposure for 1000 s and the final fluorescence intensity F was measured. Using these data, F0/F was calculated and plotted as a function of time of irradiation (t) to determine the photobleaching constant (kPB) according to the following equation:
The photobleaching constant kPB (sec-1) represents the inverse of the time to obtain 50% quenching of the initial intensity. In this way, kPB describes the entire decay, while r describes the very initial decay.
Digestion of DmsD into fragments for spectroscopy was done with 25 μg protein denatured for 20 min at 90 ᴼC prior to incubating with 0.8 μg Proteinase K overnight at 37 ᴼC in a final volume of 300 μl. Following digestion, samples were diluted to 10 μM DmsD-Trp for fluorescence spectroscopy measurements.
Statistical analyses were performed using an unpaired student t-test with 95% confidence on 3-5 experimental replicates, unless otherwise indicated. Standard error values of the mean were determined using the following equation to propagate error, using the quenching rate, r, as an example:
Mass Spectrometry
Sample digestion was performed similar to that described by Ladner et al. [27]. Native and UV-irradiated samples were heated at 90 ᴼC for 20 min prior to digestion with a 1:4 ratio of chymotrypsin-to-DmsD overnight at room temperature. The digested samples were dried using a Savant SpeedVac concentrator (Thermo Scientific) equipped with a refrigerator condensation trap, then trifluoroacetic acid (TFA) was added to a final concentration of 0.1% v/v to 100 μg/ml of digested protein.
An ÄKTA purifier HPLC (GE Biosciences) system was used for separating 100 μl of digested sample with an analytical Luna 5 μm C18(2), 250 x 4.6 mm column (Phenomenex). The mobile phase components were: A, 0.065% v/v TFA in water and B, 0.03% v/v TFA in acetonitrile. The column was equilibrated with 3 column volumes of A at 0.5 ml/min, then the digested peptides were eluted with a linear gradient of B over 10 column volumes at 1 ml/min. Peptide fragments were detected by absorption spectroscopy at 210 and 280 nm and 2 ml fractions were collected. Each fraction was vacuum dried as above and stored at -20 ᴼC until ready for MS analysis.
MALDI-TOF MS spectra were obtained on a Voyager DE-STR (Applied Biosystems) at the Southern Alberta Mass Spectrometry facility (University of Calgary) exactly as described by Ladner et al. [27]. Peaks with s/n >3 were labeled using M/Z program (Genomic solutions Inc.). Mass results are shown as mean ± standard error.
Differential Scanning Calorimetry
The heat of denaturation of DmsD was measured on a VP-DSC microcalorimeter (MicroCal Incorporated) as described by Winstone et al. [26].
RESULTS
Photobleaching Phenomena of DmsD
The native sequence of E. coli DmsD (UniProt ID P69853) contains an unusually high number of tryptophan residues (eleven), and ten tyrosine residues (Fig 1a). Upon performing routine protein fluorescence spectroscopy, considerable amount of quenching was observed. This led us to investigate the origin of photobleaching observed in DmsD. In order to appropriately compare our experiments in this study, all experimental protein concentrations indicated throughout are based on moles of Trp, and thus use the nomenclature of molar DmsD-Trp. Initial characterization of 10 μM DmsD-Trp by fluorescence spectroscopy at 280 nm excitation showed a rapid decrease of the relative fluorescence with repetitive scanning (Fig 1b). After 20 cycles of scanning (i.e. 1000 s of 280 nm irradiation), fluorescence intensity at the emission maximum (λmax) decreased by almost 70%. The fluorescence intensity decrease exhibited an exponential decay, where the initial decrease was ~8% after the first cycle, ending with ~3.5% after the 20th cycle (Fig. S1). The λmax shifted by ~4 nm over the course of 20 cycles of irradiation (Fig 1c). λmax remained stable at 348 nm for the first 400 s of irradiation but as the irradiation time increased, it showed a red shift to 350 nm after 700 s, and after 1000 s it shifted to 352 nm (Fig 1c). This wavelength shift is typical of increased solvent exposure, suggesting that the protein began to denature from the continuous irradiation.
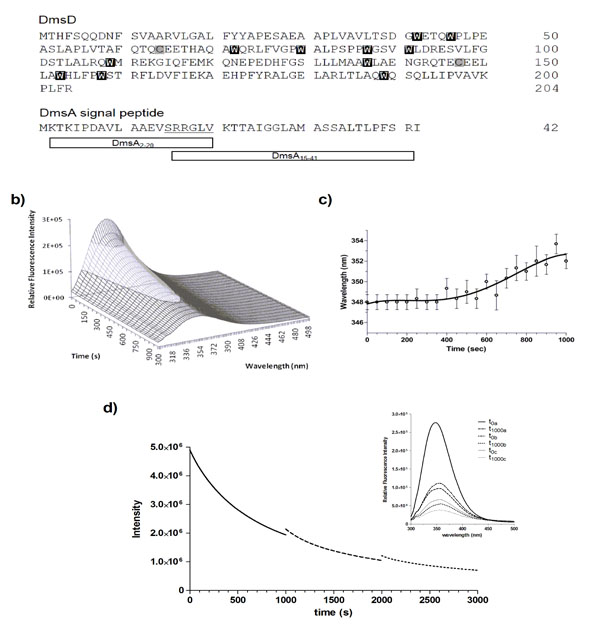
DmsD is photobleached upon repeated UV irradiation. a) Protein sequences of E. coli DmsD and DmsA leader/signal peptide. Corresponding Trp and Cys residues in DmsD are highlighted. DmsA peptides corresponding to regions that either bind or do not bind DmsD used in this study are shown. b) Fluorescence emission spectra of DmsD over 20 cycles of UV irradiation. Fluorescence intensity after the 1st and 20th scans were 2.8 ± 0.091 x 105 and 8.7 ± 0.22 x 104 counts/s, respectively. c) DmsD emission maximum (λmax) shift over the course of irradiation. d) Steady-state time-dependent decrease of DmsD fluorescence emission was recorded by irradiating for 1000 s at 280 nm (―), the sample was then removed from the fluorimeter and stored in the dark overnight at 4°C. Sample was then re-irradiated a further 1000 s (---), stored again at 4°C overnight, and the procedure repeated a third time on the same sample being irradiated for a further 1000 s (∙∙∙). Inset shows the fluorescence emission spectra of the sample. The first spectrum (t0a) was collected, then the sample was irradiated for 1000 s and then a spectrum was collected (t1000a). This protocol was repeated for each of the subsequent irradiations, providing the spectra before and after each step. The relative fluorescence intensities at various stages of irradiation and following storage in counts/s were 2.8 ± 0.27 x 105 (t0a), 9.9 ± 1.0 x104 (t1000a), 1.1 ± 0.048 x 105 (t0b), 5.5 ± 0.096 x104 (t1000b), 6.7 ± 0.043 x 105 (t0c), and 3.8 ± 0.042 x 105 (t1000c). The quenching rates were determined as ra = 1423 ± 53 s-1 for the last 100 s of the initial irradiation (a) and rb = 2494 ± 383 s-1 for the first 100 s after the first storage (b).
To determine whether the self-quenching of DmsD fluorescence was reversible or not, 10 μM DmsD-Trp solutions that were previously irradiated for 1000 s at 20 ᴼC and stored overnight at 4 ᴼC were subjected to irradiation again under the same conditions. Alignment of the three spectra shows a continuous fluorescence decay trend (Fig. 1d, inset). The initial irradiation step (t1000a) compared to prior irradiation (t0a) showed that the fluorescence intensity had decreased by ~65% and the λmax shifted to 354 nm (Fig. 1d). After storage overnight, the initial fluorescence intensity at time 0 (t0b) was slightly higher by ~11% than the intensity at t1000a suggesting that the fluorescence had recovered slightly by storage overnight (p-value = 0.0234). λmax had also blue shifted to 352 nm supporting the slight recovery. Following continuous irradiation of the sample again (t1000b), the fluorescence intensity decreased by ~50% and λmax shifted further to 356 nm. This procedure was repeated again and initial fluorescence intensity after storage overnight (t0c) recovered by ~12% (p-value = 0.0008), but the λmax remained at 356 nm. Following another cycle of irradiation, the fluorescence intensity (t1000c) decreased by ~43% and the λmax shifted further to 358 nm. It is clear from Fig. (1d) that the initial intensity of the second scan does not correspond to the final intensity following the previous UV irradiation. DmsD treated for the same period of time concurrently without irradiation showed no effect on its spectra (data not shown).
We calculated the quenching rate, r, for the first 100 s after the first storage (b) and the last 100 s of the initial scan (a) and found that the rb was ~75% higher than ra and differed significantly (p-value = 0.0087). This allows us to exclude the possibility of photo-recovery, suggesting an irreversible modification of the primary fluorophores responsible for the emission from DmsD. Therefore, the remarkably rapid loss of fluorescence from DmsD is a result of an irreversible photobleaching event upon 280 nm irradiation.
Physiochemical Conditions that Influence Photobleaching of DmsD
We explored various factors that might contribute to the photobleaching effect of DmsD. Standard experiments were performed with a relative DmsD-Trp concentration of 10 μM, therefore initial quenching rate, r, for the first 30 s of irradiation was determined for DmsD-Trp concentrations between 1 to 100 μM (Fig. 2a). From 1 to 50 μM, the quenching rate increase can be described by a second order reaction, reaching its maximum at 50 μM (r = 11367 ± 620 s-1). For 75 and 100 μM concentrations, the rate decreases to ~7500 s-1 and their difference is not significant (p-value = 0.8026). We also note that the emission spectrum of a 75 or 100 μM showed similar relative fluorescence intensity maxima (Fig. S2a; p-value = 0.0679). Decreasing to 50 μM showed a significantly lower initial intensity (Fig. S2b; p-value = 0.0027). The data in a modified Stern-Volmer plot are shown in supplementary Fig. ( S2a). Photobleaching constants (kPB) were calculated to compare if there would be any inner filter effects. For concentrations less than 10 μM, kBP values do not differ significantly (Fig S2c; p-value = 0.8689). However, at concentrations >10 μM, reduction in kBP values was observed and reached the lowest at >75 μM.
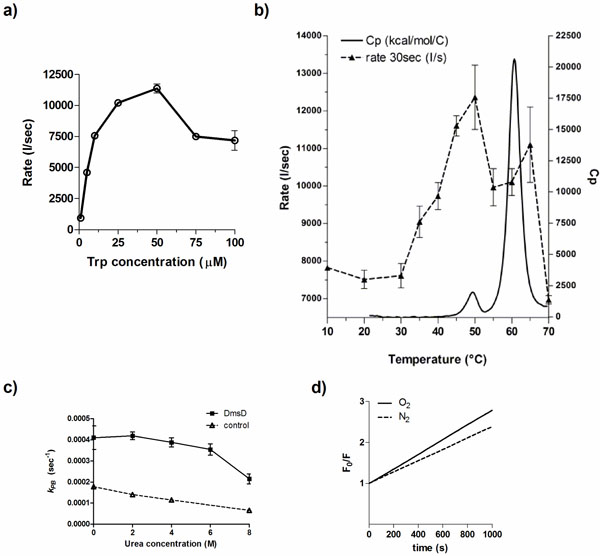
Factors that influence the photobleaching of DmsD. a) Initial quenching rates of DmsD was measured under steady state irradiation of DmsD concentrations standardized to the Trp levels in the protein from 1 to 100 µM DmsD-Trp. b) The quenching rate (---) of DmsD at different temperatures overlaid to the thermogram from differential scanning calorimetry (heat capacity, Cp; ―). Experiments were performed at the equivalent of 10 µM DmsD-Trp concentration. c) Photobleaching of DmsD and control (N-acetyltryptophanamide, NAWA) irradiated as in a) with the presence of the indicated urea concentrations. Experiments were performed at the equivalent of 10 µM DmsD-Trp or NAWA. d) Modified Stern-Volmer plot of the effect of oxygen on DmsD photobleaching rates. Samples containing 10 µM DmsD-Trp was purged with nitrogen gas (N2, ---) or air (O2, ―) prior to UV irradiation. Photobleaching constants (kPB) of nitrogen- and air-purged samples were 1.381 ± 0.1946 × 103 s-1 and 1.784 ± 0.1239 × 103 s-1, respectively.
Temperature also plays an important role in photobleaching of fluorophores [28]. To test whether DmsD photobleaching is affected by temperature, a 10 μM DmsD-Trp sample was excited at 280 nm for 1000 s at different temperatures. Between 10 to 30 °C, the quenching rate of DmsD fluorescence remained relatively similar (Fig. 2b; p-value = 0.5565). At temperatures >30 ᴼC, the quenching rate increases proportionally with temperature and reaches a maximum at 50 ᴼC. The change in quenching rate at this temperature corresponds to a pre-transition unfolding state of the protein as evidenced by differential scanning calorimetry (Fig. 2b). This may be due to a conformational change in DmsD that exposes partially buried tryptophan residues. At temperatures >50 ᴼC, the quenching rate rapidly decreases with the protein becoming unfolded at above 60 ᴼC. Additionally the λmax shifted according to temperature changes, initially at 346-348 nm to 352-358 nm with unfolding as expected from exposure of the Trp to the aqueous environment (data not shown). The modified Stern-Volmer plot for temperatures between 10 to 30°C is linear and photobleaching constant kPB increases with temperature (Fig. S3a). Starting from 35°C, the plot shows a downward curvature (Fig. S3b- d) and the kPB increase was linear until 50°C, and then it decreased from (Fig. S3e). Overall the data here correlate well with thermal unfolding and demonstrate the observed irreversible photobleaching is a result of folding within DmsD.
To further test whether protein unfolding had an effect on photobleaching, DmsD in varying concentrations of the denaturant urea was irradiated. At concentrations up to 6 M urea, the difference between photobleaching constants was non-significant (Fig. 2c; p-value = 0.5105, One-Way Analysis of Variance, ANOVA). However, inclusion of the photobleaching constant at 8 M urea for analysis indicates that the sum of means had significant variation (Fig. 2c; p-value = 0.0157). At this concentration of urea, DmsD is expected to be completely unfolded [8], similar to the effect observed during thermal unfolding in Fig (2b). Both observations suggest that unfolded DmsD exhibits considerably reduced photobleaching demonstrating that the phenomenon is conformation dependent.
The photobleaching effect was not solely due to the presence of oxygen, a known fluorescence quencher and tryptophan photobleacher [29]. Samples purged with nitrogen gas showed that the initial fluorescence intensity did not differ significantly from a sample purged with air (Fig 2d; p-value = 0.5675). During 280 nm irradiation, quenching still occurred regardless of the presence of oxygen, although kPB differed significantly under the nitrogen-purged conditions (p-value = 0.0391). This difference suggests that a small portion of the photobleaching is catalyzed by the oxygen, likely to the indole derivative N-formylkynurenine (NFK).
Conformational Changes Influence Photobleaching of DmsD
DmsD has been shown to have multiple folding forms on native acrylamide gels as a result of exposure of differential amounts of charge [8]. The cysteine reducing agent dithiothreitol (DTT) is added to DmsD during purification as it appears that DmsD is trapped in a specific folding variant form under cysteine oxidizing conditions [8]. Without DTT, the stability of DmsD in solution is also decreased (Winstone et al, unpublished). To assess whether the presence of DTT affects photobleaching of DmsD, DTT was removed by dialysis just prior to experimentation. In the presence of DTT, the photobleaching constant of DmsD is lower, suggesting that it has a protective effect against photobleaching (Fig. 3a). This observation is prevalent at all temperatures tested between 20 to 70 ᴼC, and the gap between the two samples (with and without DTT) remains nearly the same. As DTT can also affect the folding of DmsD, it is plausible that it has a dual effect in protection and through stabilization of its folding. The presence of DTT did not appear to shift λmax but did induce the formation of an additional peak with λmax at ~430 nm (Fig. 3b). At this emission maximum, a possible product is NFK (λmax 435 nm; [30]) or an unidentified product previously described with λmax at 433 nm and biochemically distinct from NFK [31]. This new fluorescent peak suggests that excited state chemistry has occurred, catalyzed by free cysteine thiols with the indole ring generating a new chromophore, which would be consistent with photoproduct of α-crystallin that is enhanced with glutathione, upon 300 nm irradiation [31]. At the observed red shifted emission, the product is unlikely ditryptophan or similar derivatives, whose emission maxima are between 350 to 400 nm [32]. However, other studies by our group have shown that the addition of various R-group modifications to the indole ring leads to red shifted fluorescence [33]. This data along with the folding variant observation published in the work of Sarfo et al. [7] and Winstone et al. [8] suggests that the folding form without DTT leads the protein to be in a conformation that orientates the Trp residues in an environment that increases photobleaching kinetics.
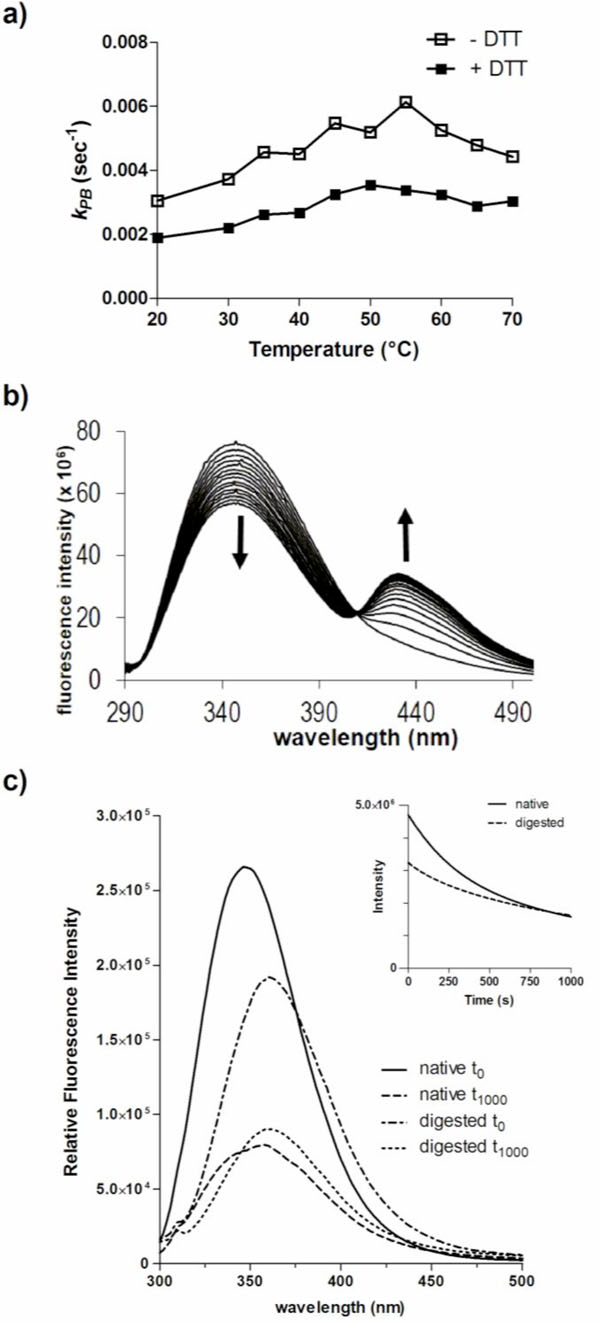
Photobleaching of DmsD is influenced by its conformation. a) Effect of the reducing agent DTT on photobleaching of DmsD. The photobleaching constant (kPB) of 10 µM DmsD-Trp was determined between 20 to 70 °C in the presence or absence of 1 mM DTT. b) DTT induces a new bimodal peak in the fluorescence spectra of DmsD. c) Emission spectra of native DmsD and proteinase K digested DmsD collected prior to (t0) or after UV irradiation (t1000). Relative fluorescence intensities in counts/s of native samples at λmax were 2.6 ± 0.25 x 104 (t0) and 7.9 ± 0.89 x 103 (t1000), and of digested samples were 1.9 ± 0.079 x 105 (t0) and 9.0 ± 0.48 x 104 (t1000). Quenching rates (r) of undigested and digested samples were 7644 ± 197 s-1 and 4078 ± 151 s-1 respectively.
To further assess the effects of conformation on photobleaching, DmsD was digested with proteinase K into peptides prior to 280 nm irradiation. Since DmsD was present in ~400-fold excess (in moles) compared to proteinase K, the protease was not removed prior to spectroscopy measurements as its contribution would be negligible. Proteinase K cleaves adjacent to aliphatic and aromatic residues therefore we predict that each peptide would contain on average approximately one Trp residue. The entire emission spectra before and after irradiation were collected and compared to native undigested DmsD. A significant decrease in relative fluorescence intensity (p-value = 0.0175) and a λ max shift was observed between the native and the digested samples (Fig. 3c). The initial relative fluorescence maximum of the digested sample at 348 nm was ~73% relative to that of the native undigested sample at 360 nm. After irradiation, native DmsD relative fluorescence decreased by ~70% and λ max red shifted by 10 nm as observed above. The relative fluorescence of digested sample decreased by ~52% and a red shift of λ max by only 2 nm, although digested DmsD was already red shifted compared to undigested, demonstrating the Trp residues were mostly solvent exposed after digestion. The photo-quenching rate, r, of the undigested sample was almost double of the digested sample and differed significantly (p-value < 0.0001).
Photobleaching of native DmsD was much higher than when digested, as judged by the nearly twice higher kPB value (Fig. S4; p-value = 0.0037). The decreased fluorescence of the digested sample compared to the native protein is likely due to a loss of energy transfer between contiguous residues as well as an increase in dynamic freedom of the chromophores. Again, digested DmsD was already red shifted compared to undigested.
Identification of Photoreacted Chromophores
To determine which Trp residues were being chemically damaged resulting in the photobleaching, irradiated and non-irradiated native DmsD was subjected to proteolysis and the resulting peptides were analyzed by Matrix-assisted laser desorption ionization mass spectrometry (MALDI-TOF MS). Samples were first digested with chymotrypsin into peptides and separated by HPLC. Majority of the peptides eluted at a retention time between 16 to 33 min but the greatest differences was observed between 16 to 24 min (Fig. S5). Therefore fractions collected from these retention times were selected to be analyzed by MALDI-TOF MS. Supplementary Table S1 lists the characteristics of identified peptide fragments either in non-irradiated or 280 nm irradiated DmsD protein samples. Note that not all peptides contained Trp residue as chymotrypsin preferentially cleaves after Phe, Tyr, and Trp residues [34]. Three Trp residues were identified with a molecular weight difference corresponding to an oxygen modification – W42, W80, and W109 (Table S1). Residues W42 and W80 appeared to have an added mass corresponding to one oxygen molecule whereas W109 appeared to have two. It has been reported that addition of up to four oxygen molecules can occur on Trp upon UV irradiation [35]. With only a single oxygen molecule added to W42 and W80, these likely correspond to the formation of oxindolylalanine or hydroxytryptophan [36]. W109 contained the added mass of two oxygen molecules, suggesting the formation of di-oxindolylalanine or NFK. All of the four derivatives are known products previously described to be oxidation products of tryptophan [36-38]. MS profiling experiments found that these four and dihydroxytryptophan are potential products generated from photo-oxidative degradation of tryptophan corresponding to the addition of one or two oxygen molecules, supporting that these are the resulting Trp derivatives detected in DmsD peptides [39].
Mapping of these residues onto the E. coli DmsD crystal structure, 3EFP.pdb [18], showed that W42 is in a mobile loop that is partially exposed, W80 is located on the surface and is completely exposed, and W109 is also partially exposed (Fig. 4a & b). We found that amongst all the identified peptide fragments, those consisting of residues 81-104, 138-162, and 159-174 were missing from the irradiated sample (Supplementary Table S1). Changing experimental conditions failed to produce these peptides in the mass profile. There are several Trp residues in these missing fragments which include W87, W91, W137, and W158. Residues W87 and W137 are partially exposed in the structure, whereas W91 and W158 are buried (Fig. 4c). Focusing on these residues within the structure shows that W91 and W158 face each other and are 3.5 (C2Trp91-C6Trp158) to 7.5 Å (C6Trp91-C6Trp158) apart, suggesting that they could interact (Fig. 4d).
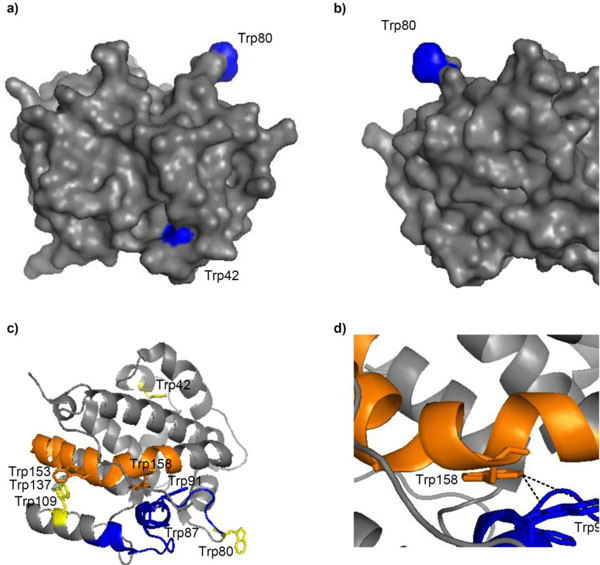
Trp modifications in E. coli DmsD (3EFP.pdb) after UV irradiation. a & b) Surface localization of residues W42, W80 and W109; images are a 180° rotation about the y-axis. c) Cartoon representation of DmsD with peptide fragments 81-104 (blue) and 138-162 (orange) that were not identified by MALDI-TOF MS analysis from the UV irradiated sample. Each peptide is preceded by a Trp, a putative cleavage site of chymotrypsin. Residues W42, W80, and W109 are colored yellow for comparison. d) Expanded view of c) showing the proximity of W91 and W158.
W87 is a residue conserved in DmsD homologs and is found within the binding pocket of DmsD towards its substrate DmsA RR-signal peptide [20]. It appears to be partially exposed in the final structure and could be a good candidate to understand the photobleaching differences. Using a W87S mutant variant of DmsD, we found that the final fluorescence intensity after UV irradiation was ~20% higher than wildtype DmsD (Fig. 5a). The initial quenching rate of W87S variant was ~40% lower compared to wildtype (Fig. 5b), suggesting that this residue is an important contributor to the total fluorescence and its loss is noted in the photobleaching. This observation is also supported by the reduced kPB values for the mutant (Fig. 5c). According to these data, it seems that the region containing W80, W87, and W91 (through interactions with W158) is conformational key to the photobleaching phenomena of DmsD.
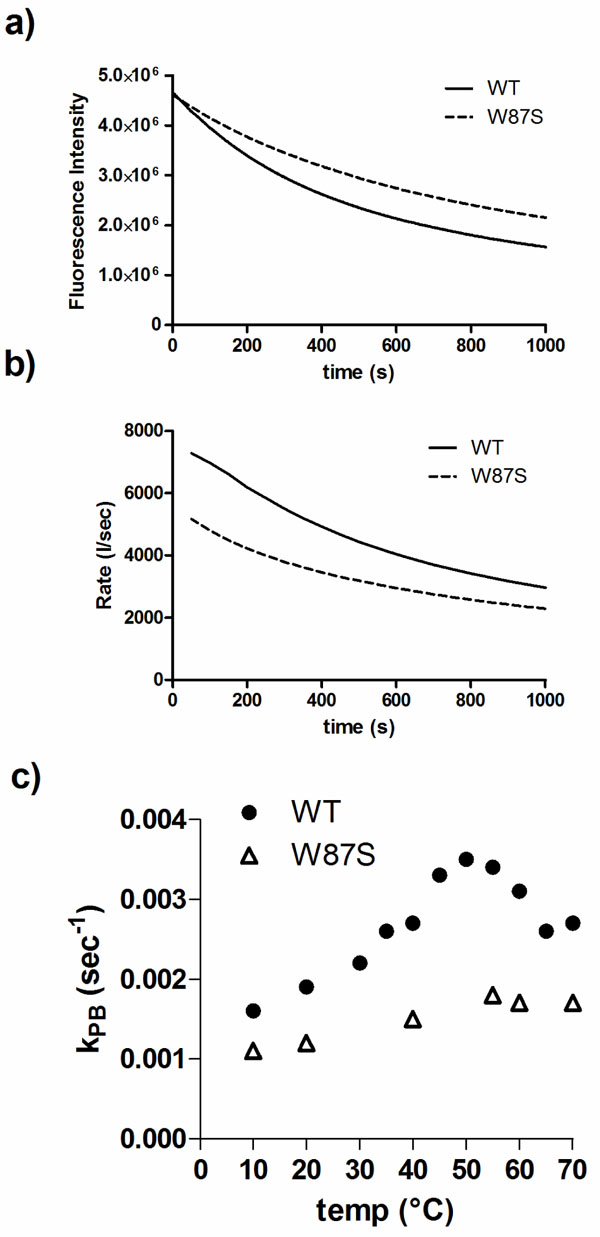
Trp87 is a significant contributor to photobleaching of DmsD. a) Fluorescence intensity changes of a W87S DmsD mutant compared to wildtype (WT) upon 280 nm irradiation. b) Comparison of the self-quenching rate of WT and W87S DmsD of samples from panel a. c) Quenching of WT and W87S DmsD mutant at various temperatures with photobleaching constants (kPB) indicated.
Effect of DmsA Signal Peptide Binding to DmsD on Photobleaching
The RR-signal peptide of DmsA is a well-defined substrate of DmsD that binds in the micromolar range [6,8,9,14,20]. Molecular dynamic simulations with DmsD found that it undergoes conformational adjustments upon binding to the signal peptide [18]. The sequence of the RR-signal peptide can be divided into two regions that differ in binding to DmsD (Fig. 1a) – residues 2-20 (DmsA2-20) do not bind and residues 15-41 (DmsA15-41) interact well [26]. We studied whether the addition of these peptides to DmsD prior to irradiation has any effect on photobleaching. According to the simulation results as described in the work of Stevens et al. [18] and circular dichroism spectroscopy in Winstone et al. [26], DmsA RR-peptide binding should induce a conformation change in DmsD and thus may alter its photobleaching. We found that the presence of peptide DmsA15-41 resulted in a significant increase of the photobleaching constant (Table 1; p-value = 0.0015), while DmsA2-20, had no significant effect (p-value = 0.7561). The sequence of both DmsA peptides does not contain any Trp (or Tyr) residues and thus did not contribute to the observed effects. This effect was also evident when comparing the fluorescence intensity in a modified Stern-Volmer plot (Fig. S6), where the F0/F ratio after 1000s of irradiation was ~10% higher in the presence of DmsA15-41 peptide. The results suggest that binding of the DmsA leader peptide affects the conformation of DmsD in a way which can alter the orientation of the tryptophan residues to increase photobleaching.
Effect of DmsA Signal Peptide on the Photobleaching of DmsD. Photobleaching Constant (kPB) was Calculated based on the Equation Described in the Methods section. The Dissociation Constant (Kd) of DmsA Signal Peptide Binding was Determined by Isothermal Titration Calorimetry from Winstone et al [26].
| Signal peptide added | kPB (×10-3 s-1) | Kd(µM) |
|---|---|---|
| None | 1.117 ± 0.0295 | n/a |
| DmsA2-20 | 1.156 ± 0.0311 | NB1 |
| DmsA15-41 | 1.387 ± 0.0111 | 1.7 ± 0.2 |
1 Binding not detected.
DISCUSSION
Irreversible photobleaching of DmsD by 280 nm irradiation appears to be a unique phenomenon related to its conformation and folding state. Tryptophan residues can undergo photo-oxidative modification and degradation [39], and the data observed here suggest that Trp residues in DmsD are undergoing both photo-oxidation and a conformational dependent photobleaching. Photo-oxidative degradation of Trp-containing peptides was previously observed in peptides irradiated for 12 hours [39], yet irreversible degradation of Trp in DmsD occurred in the span of ~16 minutes, a typical timeframe in which many fluorescence spectroscopy experiments are performed. This suggests that DmsD contains a unique conformation and dynamics leading to the observed rapid photobleaching phenomena.
DmsD contains 11 tryptophan residues, equivalent to ~6% of its sequence composition. Considering that the average composition is ~1% Trp, as per comparison to the human proteome with a bank of 37,866 identified proteins with an average of 1.2% Trp [40], this is highly unusual. At the excitation wavelength to which our experiments were conducted, it is possible for energy transfer from Tyr to Trp to occur [41]. Given that we observed a decrease in Trp fluorescence upon UV excitation, should energy transfer occur between these residues, the result would be an increase in fluorescence, suggesting that this unique observation is due to other factors that cause fluorescence quenching.
The data presented here support that photobleaching can be affected by folding and conformation changes in DmsD. The quenching rate increased with temperature possibly due to increased exposure of mobile loops containing Trp residues to the quenching elements. When the protein was fully denatured by high temperature due to thermal unfolding, quenching was drastically reduced. There appeared to be no urea concentration-dependent effects on the quenching rate (not shown); rather a significant decrease in quenching efficiency as evidenced by a reduced photobleaching constant in the presence of 8 M urea. DmsD loses ~50% of its ability to bind DmsA RR-signal peptide between 3-4 M urea and at 8 M almost no binding is observed [8]. These observations suggest that self-quenching and subsequent photobleaching of DmsD are due to its folded conformation, also supported by the recovery of fluorescence upon overnight storage of the protein. Of the three Trp residues resulting in oxygen modifications upon UV irradiation, W87 was previously shown to be important for DmsA RR-signal peptide binding [18,20]. The previously identified DmsA signal peptide binding pocket also includes W72, W80, and W91, amongst other non-Trp residues. W91 is in close proximity with W158 and we hypothesize that it contributes to the photobleaching phenomena through interactions with W158. By targeting W87, a conserved residue amongst DmsD close homologs [20], we found that it was indeed a major contributor to photobleaching of DmsD, an effect that can be adverted by simply mutating it to serine. In fact, the photobleaching constant of the mutant appeared to be mostly unaffected by thermal denaturation unlike wildtype DmsD (Fig. S4).
It has been demonstrated that a red-shift of Trp occurs when the environment becomes more polar, and fluorescence of Trp is dependent on neighboring amino acids in the sequence [42]. The magnitude of photobleaching in DmsD is unprecedented, with the exception of human interferon-α2a whose photobleaching decay is 18 min [43]. Thus along with the observation of multiple folding forms of this protein [7], this protein’s high Trp concentration and unique conformational environments around the fluorophore indicate its uniqueness in this regard.
The extensive photobleaching that DmsD experiences would limit the use of fluorescence spectroscopy to study it, yet the experiments here evaluating photobleaching demonstrate that this effect is very sensitive to the conformational state of DmsD. Binding of the substrate RR-signal peptide to DmsD induced a conformational change to increase its photobleaching. This is supported by observations that buried residues in DmsD become more surface exposed upon docking of the DmsA signal peptide by docking and molecular dynamics studies [18]. Residue W72 was shown to form van der Waals interactions with the signal peptide, whereas W80 and W91 formed hydrogen bonds. The binding of the DmsA RR-leader peptide to DmsD has been a challenge to understand. Although the interaction is in the affinity level of 0.5-2 µM, it has been difficult to observe any conformational changes involved. Thus we have managed to utilize this unique phenomenon to serve as a sensor of the subtle conformational changes involved with RR-leader binding.
CONCLUSION
Here we describe a unique photobleaching phenomenon in DmsD that is affected by its folding and conformation. Upon further investigation, we found that the key contributors to this phenomenon involve W87, a residue that is found within the DmsA leader binding site. Additional Trp residues involved in DmsA leader binding may also be involved as binding affects the phenomenon. We suggest that the photobleaching phenomenon in proteins should not hinder one’s use of fluorescence spectroscopy to study the protein of interest.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material can be viewed at Download Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. R.A. Edwards for useful discussions. This research was funded by a Canadian Institute of Health Research operation grant to RJT.


