All published articles of this journal are available on ScienceDirect.
Histidine 117 in the His-Gly-Ser-Asp motif is Required for the Biochemical Activities of Nucleoside Diphosphate Kinase of Mycobacterium smegmatis
Abstract
Nucleoside diphosphate kinase (NDK), which is widely conserved in both prokaryotes and eukaryotes, maintains a balanced pool of nucleotide triphosphates and their deoxy derivatives. NDKs from bacterial and other systems contain the conserved HGSD motif, where the His residue is required for the biochemical activities, namely the NTPase (AT-Pase and GTPase), NTP synthesising, and autophosphorylation activities of the enzyme. Amino acid sequence homology comparison of the NDK of Mycobacterium smegmatis (MsmNDK) with the NDKs of other bacterial genera showed the presence of H117GSD motif. While the recombinant wild type MsmNDK showed the NTPase, NTP synthesising, and autophosphorylation activities, the H117Q mutation abolished the biochemical activities of the recombinant MsmNDK-H117Q mutant protein in vitro. These observations demonstrate that the H117 residue in the HGSD motif is required for the biochemical activities of MsmNDK.
INTRODUCTION
Maintenance of the level of nucleoside triphosphates (NTPs) as well as their corresponding deoxy derivatives (dNTPs) is crucial to all growth and developmental processes. The enzyme nucleoside diphosphate kinase (NDK), first discovered simultaneously by Krebs and Hems [1], and Berg and Joklik [2], utilises an autophosporylated enzyme to mediate the transfer of 5’ terminal phosphate from NTPs (mostly ATP) to nucleoside diphosphates (NDPs or their deoxy derivatives), to generate the respective NTPs, via a reversible mechanism, as given below [3-7].
N1TP + N2DP ↔ N1DP + N2TP
Thus, the mechanism implies that NDK is a substrate non-specific enzyme [4-8], which can remove the gamma phosphate from ATP or GTP to show ATPase and GTPase activities, transfer the removed phosphate to NDPs to generate NTPs (NTP synthesising activity), through the phosphorylation of the His residue in the conserved HGSD motif (autophosphorylation activity) on the enzyme [8-10].
The primary function of NDK is to maintain NTP and dNTP concentrations, as NDK deficiency in Escherichia coli leads to nucleotide pool perturbation [4, 11], resulting in the incorporation of inappropriate nucleotides into DNA [11, 12]. E. coli NDK was also found to possess uracil-DNA glycosylase, AP-lyase, and 3'-phosphodiesterase activities in vitro [13]. Similarly, the human NDK homologues, NDK1, NDK5, NDK7 and NDK8 proteins, were found to possess 3’-5’ exonuclease activity [14]. It has a role in the growth and differentiation in Myxococcus xanthus [15] and Pseudomonas aeruginosa [9], and in tumor metastasis suppression in humans [16]. NDK in association with succinyl CoA synthetase in P. aeruginosa [17] and with pyruvate kinase, EF-Tu, and a cell wall protein in Mycobacterium smegmatis [18] synthesises specific NTP, required for the activity of the associated protein. In Drosophila melanogaster, NDK is involved in wing disc development [19]. The ATPase activity of the secreted form of Mycobacterium tuberculosis NDK (MtuNDK) causes cytotoxicity to macrophages [20]. MtuNDK was found to block phagosome maturation in murine Raw 264.7 macrophages [21]. In the present study, as the first step towards understanding the physiological roles of NDK in mycobacteria, we present evidence that the His residue in the His-Gly-Ser-Asp is required for all the biochemical activities of Mycobacterium smegmatis NDK in vitro.
MATERIALS AND METHODOLOGY
Bacterial Strains and Culture Media
E. coli JM109 and E. coli M15 cells were used for the propagation of recombinant clones and for the overexpression of protein, respectively. Luria broth with ampicillin (100 μg/ml) or kanamycin (25 μg/ml) was used for the selection and culturing of recombinant clones.
Cloning of Msmndk Gene
Full-length Msmndk gene (MSMEG_4627) was cloned in pBS(KS) vector between KpnI and EcoRI sites, after PCR amplification from M. smegmatis mc2155 genomic DNA using Msndk-f2 and Msndk-r2 primers (Table 1). The resultant pBS-Msmndk clone was sequence verified. The Msmndk ORF was then subcloned into pQE30 expression vector between KpnI and PstI sites, to obtain pQE30-Msmndk construct.
Oligonucleotide Primers Used
| Name of the Oligo Primer | Sequence of the Oligo Primer | For Cloning |
|---|---|---|
|
|
||
| Msndk-f2 | cggggtaccgtgactgagcggaccctcgtacttatcaagcc | MsNDK ORF cloning |
| Msndk-r2 | ccggaattcggcggtggcctcgccggg | MsNDK ORF cloning |
| MsNDK-H117Qf | cacgcaggacaatctcgtgcagggttccgattc | MsNDK-H117Q cloning |
| MsNDK-H117Qr | ctcgggcgaatcggaaccgtccacgagattg | MsNDK-H117Q cloning |
Note: Restriction enzyme sites are underlined. The mutation introduced is given in bold nucleotide letters.
Generation of MsmNDK-H117Q
The MsmNDK-H117Q mutant was generated by overlap extension PCR, using Msndk ORF-specific primers and Msmndk mutagenic primers (Table 1). Using ORF-specific forward primer (Msndk-f2) and mutagenic reverse primer (MsNDK-H117Qr) a 360 bp amplicon and using the forward primer (MsNDK-H117Qf) and ORF-specific reverse primer (Msndk-r2), another product of 60 bp were generated. These two mega primers were again used for the PCR reaction under the same condition for 5 cycles to generate megaprimers. Subsequently, the ORF-specific forward (Msndk-f2) and reverse primer (Msndk-r2) were introduced into the reaction mixture. The PCR reaction was then continued for 25 more cycles, and the PCR product was cloned in pBS-KS between KpnI and EcoRI sites, to get pBS(KS)-MsmNDK-H117Q. The mutation was confirmed using DNA sequencing. The mutant Msmndk ORF was then subcloned into pQE30, between KpnI and PstI sites, to generate pQE30-MsmNDK-H117Q mutant clone. Both the recombinant proteins 6xHis-MsmNDK and 6xHis-MsmNDK-H117Q were overexpressed and purified from soluble fraction to near homogeneity using Ni2+-NTA agarose affinity chromatography. The protein preparations were dialysed against lysis buffer containing 10% glycerol. Dialysis was carried out at 4ºC with three changes of 500 ml buffer to remove imidazole and other salts, and the protein was stored at -75ºC.
Circular Dichroism (CD) Spectroscopy
Circular Dichroism spectroscopy experiments were carried out using spectropolarimeter (JASCO J-715) measuring CD values over 200-260 nm wavelength range at every 0.5 nm interval, in 2 mm path-length cuvette. A concentration of 8.93 µM protein (4.2 mg/ml) was used in 1 ml of 1 mM Tris-HCl (pH 8.0) and 5 mM KCl buffer, per assay at room temperature. The CD values were processed and plotted as graph using Microsoft Excel 2003. CD spectrum was expressed in terms of mean residual ellipticity [22].
ATPase and GTPase Assays
ATPase and GTPase assays were carried out, as described [8]. In brief, 1 µg of NDK was incubated at 25°C in 20 μl of TMD buffer (50 mM Tris-HCl, 10 mM MgCl2 and 1 mM DTT, pH 7.4) with 0.1 µCi of [γ-32P]-ATP (6000 Ci/mmol) or [α-32P]-GTP (3000 Ci/mmol) for 15 min or 30 min. The reaction was stopped with a final concentration of 2% SDS and 1 mM EDTA. The samples were heated at 75°C for 5 min, one μl of the reaction mixture was spotted on a polyethyleneimine cellulose thin layer chromatography plate (PEI-TLC, Merck), and resolved using 0.75 M KH2PO4, as described [23]. The radioactive spots corresponding to [γ-32P]-ATP and 32Pi or of [α-32P]-GTP and [α-32P]-GDP were then visualised using Phosphorimager BioImage Analyser (FLA 2000, Fuji, Japan). The regions corresponding to 32Pi or [α-32P]-GDP on TLC were cut and counted in liquid scintillation counter. The counts per minute (CPM) values were plotted as bar graph, with standard deviation.
Autophosphorylation Assay
Autophosphorylation activity of the purified wild type MsmNDK and mutant MsmNDK-H117Q proteins was measured, as described [17]. In brief, 1 µg each of the purified proteins was incubated with 0.1 µCi [γ-32P]-ATP (6000 Ci/mmol) in 20 µl of TMD buffer (50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, and 1 mm of dithiothreitol). The reaction was terminated after 10 min with 2 µl of 10% SDS. The samples were boiled for 10 min, fractionated on 12% SDS-PAGE, and analysed using autoradiography. The radioactivity in the protein bands in the dried gel was quantitated using liquid scintillation counter. The counts per minute (CPM) values were plotted as bar graph, with standard deviation.
NTP Synthesising Activity
NTP synthesising activity of MsmNDK was determined, as described [18]. Assay was carried out in 20 μl of TMD buffer containing, 1 µg of MsmNDK or MsmNDK-H117Q, 250 µM each of CDP and GDP. The reaction was initiated with the addition of 0.1 µCi [γ-32P]-ATP (6000 Ci/mmol), along with of 125 µM non-radioactive ATP. After incubation at 30°C for 10 min, the reaction was stopped with final concentration of 2% SDS and 1 mM EDTA. The samples were heated at 75°C for 5 min, and one μl each of the reaction mixture was analysed on polyethyleneimine cellulose thin layer chromatography (PEI-TLC) plates, in 0.75 M KH2PO4, as described [23]. The positions of ATP, CTP, and GTP were identified using the Rf values for these NTPs in the solvent system. The radioactive GTP or CTP were then visualised using Phosphorimager Bioimage Analyser (FLA 2000, Fuji, Japan). The radioactive spot corresponding to the unhydrolysed γ-32P-ATP was cut and counted in both cases. The counts per minute (CPM) values were plotted as bar graph with standard deviation.
RESULTS
Conservation of His-Gly-Ser-Asp Motif in MsmNDK
All the NDKs, including those of Mycobacterium tuberculosis and Mycobacterium leprae, contain the conserved motif, His-Gly-Ser-Asp (HGSD). The activity of hitherto biochemically characterised NDKs was known to be dependent on the His residue in the HGSD motif [5, 6, 20, 24-29]. Homology comparison of MsmNDK protein sequence with the amino acid sequence of NDKs of a variety of bacterial genera and other organisms showed the presence of H117GSD (Fig. 1). The possibility of the involvement of H117 residue in the biochemical activities of MsmNDK was examined using the wild type and the mutant, MsmNDK-H117Q, proteins (Fig. 2A, B). Far-UV CD spectroscopy did not show any gross alteration in the overall secondary structure of the mutant (Fig. 2C).

Multiple sequence alignment of NDK proteins from diverse bacterial genera (Invariant amino acid residues HGSD is highlighted and active site H is indicated in bold letter). Multiple alignments were carried out using clustalW (http://www.ebi.ac.uk/clustalw). EcN:NDK from Escherichia coli, PaN: NDK from Pseudomonas aeruginosa, MxN: NDK from Myxococcus xanthus, MlN: NDK from Mycobacterium leprae, MtN: NDK from Mycobacterium tuberculosis (H37Rv), MsN: NDK from Mycobacterium smegmatis (mc2155), DdN: NDK from Dictyostelium discoideum.
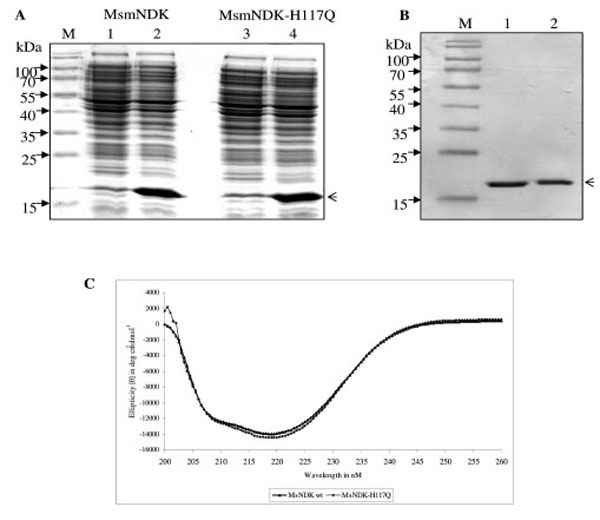
Overexpression and purification profile of MsmNDK and MsmNDK-H117Q. A. Coomassie blue stained SDS-PAGE profile of overexpressed MsmNDK (2) and MsmNDK-H117Q (4). Lanes 1 and 3, respective uninduced samples. Arrow head indicates the overexpressed or purified protein. B. SDS–PAGE of recombinant purified MsmNDK wild type (1) and MsmNDK-H117Q (2) proteins; lane M represents molecular weight markers. C. Far-UV circular dichroism (CD) spectrum of MsmNDK and MsmNDK-H117Q.
ATPase, GTPase, and NTP Synthesising Activities
The ATPase and GTPase activities of the MsmNDK and MsmNDK-H117Q proteins were determined from the hydrolysis of γ32P-ATP and α32P-GTP. Quantitation of the 32Pi released from γ32P-ATP and the α32P-GDP formed from α32P-GTP by wild type MsmNDK and MsmNDK-H117Q, from the TLC profile, showed ATPase (Fig. 3A, B) and GTPase (Fig. 4A, B) activities by MsmNDK. In contrast, very low quantities of 32Pi was released by MsNDK-H117Q, from γ32P-ATP (Fig. 3A, B), and α32P-GDP from α32P-GTP (Fig. 4A, B), as compared to that by the wild type protein. The H117Q mutation abolished the ATPase and GTPase activities to almost 4.75- and 2-fold, respectively. These observations confirmed that H117 is an essential residue required for the ATPase and GTPase activities of MsmNDK.
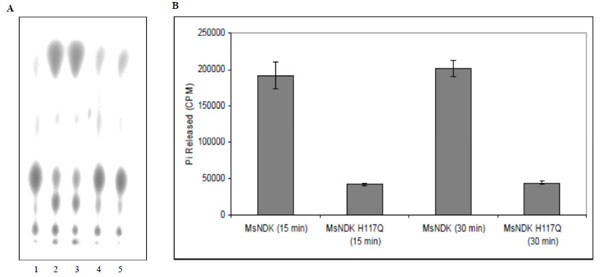
ATPase activity of MsmNDK and MsmNDK-H117Q. A. Lane 1. ATP alone, Lane 2. MsmNDK at 15 min, Lane 3. MsmNDK at 30 min, Lane 4. MsmNDK-H117Q at 15 min, Lane 5. MsmNDK-H117Q at 30 min. B. Bar graph showing the ATPase activity of MsmNDK or MsmNDK-H117Q at 15 or 30 min. Assay was repeated independently three times for each sample. Counts per minute (CPM) were recorded and standard deviations were calculated.
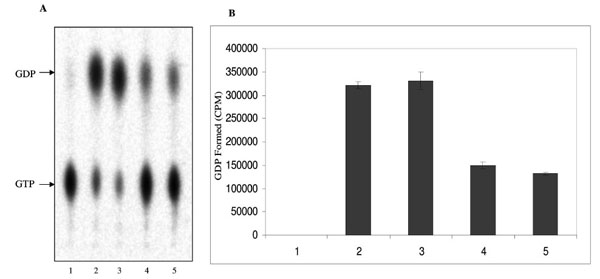
GTPase activity of MsmNDK and MsmNDK-H117Q. A. Lane 1. GTP alone, Lane 2. MsmNDK at 15 min, Lane 3. MsmNDK at 30 min, Lane 4. MsmNDK-H117Q at 15 min, Lane 5. MsmNDK-H117Q at 30 min. B. Bar graph showing the GTPase activity in terms of α32PGDP formed by MsmNDK or MsmNDK-H117Q mutant at 15 or 30 min. Assay was repeated independently three times for each sample. Counts per minute (CPM) were recorded and standard deviations were calculated.
In the NTP synthesising activity, MsmNDK converted both GDP and CDP to GTP and CTP, respectively, using the 5’ terminal phosphate from γ-32P-ATP (Fig. 5A). However, MsmNDK-H117Q showed negligible extent of conversion of GDP to GTP and CDP to CTP (Fig. 5A), showing that the H117Q mutation affected the activity (Fig. 5B). The almost complete abolition of ATP and GTP synthesis by the mutant indicated that H117 residue is essential for the NTP-synthesising activity.
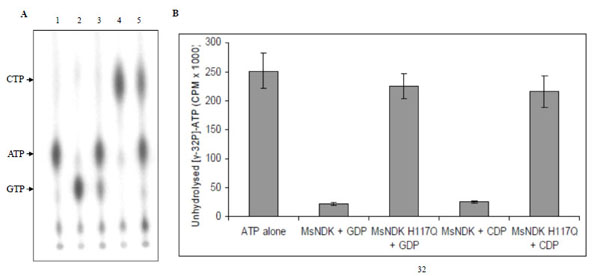
NTP synthesising activity of MsmNDK and MsmNDK-H117Q. A. PEI-TLC profile of [γ-32P]-ATP alone (lane 1), and in the presence of [γ-32P]-ATP: GDP + MsmNDK (lane 2), GDP + MsmNDK-H117Q (lane 3), CDP + MsmNDK (lane 4), and CDP + MsmNDK-H117Q (lane 5). B. Bar graph representing quantitation of unhydrolysed [γ-32P]-ATP. The positions of ATP, CTP, and GTP were identified using the Rf values for these NTPs in the solvent system, 0.75M KH2PO4. Assay was repeated independently three times for each sample. Counts per minute (CPM) were recorded and standard deviations were calculated.
Autophosphorylation Activity
While transferring the γ-phosphate from ATP to an acceptor NDP, NDK, gets autophosphorylated at the His residue in the HGSD motif [25, 28, 30]. In order to verify whether the formation of autophosphorylated intermediate of MsmNDK is through H117, recombinant MsmNDK and MsmNDK-H117Q proteins, subsequent to incubation with [γ-32P]-ATP in the absence of NDP, were run on 12% SDS-PAGE and analysed using autoradiography. A 15 kDa phosphorylated band corresponding to MsmNDK was observed, indicating that MsmNDK undergoes autophosphorylation (Fig. 6B). However, the mutation H117Q reduced the autophos-phorylation activity, almost 5-fold, indicating that H117 is involved in the autophosphorylation (Fig. 6C; protein bands inFig. 6A).
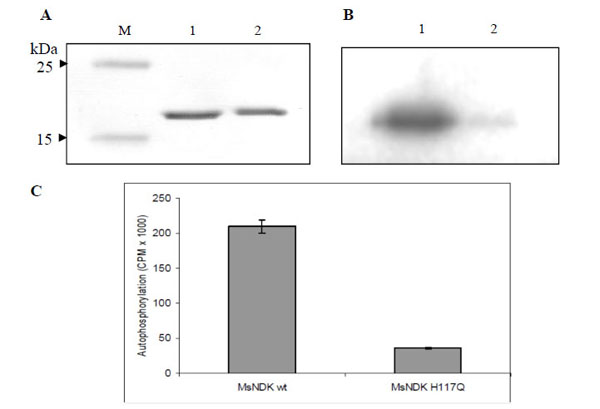
Autophosphorylation assay of MsmNDK and MsmNDK-H117Q. A. Coommassie blue stained SDS-PAGE gel profile. Lane M, Mol Wt markers. Lane 1, MsmNDK (1 µg). Lane 2, MsmNDK-H117Q (1 µg). B. Autoradiograph of MsmNDK (lane 1) and MsmNDK-H117Q (lane 2) after autophosphorylation. C. Bar graph represents the quantitation of autophosphorylation of MsmNDK and MsmNDK-H117Q. Assay was repeated independently three times for each sample. Counts per minute (CPM) were recorded and standard deviations were calculated.
DISCUSSION
The present study demonstrates that MsmNDK, which is also conserved like other structurally and functionally characterised NDKs [9, 15, 18-20, 24-26, 31-41], possesses NTPase, NTP synthesising, and autophosphorylation activities. In the attempt to find out the requirement of H117 for the biochemical activities, the intention was to find out whether even the subtle change of His to Gln would be able to affect the activities. While Gln is neutral and polar, His is basic and polar. The dipole of the C-N bond in the amide group in Gln is stronger than the dipole in the C-N bond in His. The polarity scale of His is (-) 0.49 ± 0.15, while that of Gln is (+) 0.73 ± 0.15 [42, 43]. Further, the average volume of the side chains of Gln and His [44, 45], their relative hydrophilicity, and the net charge of their side chains, are comparable [46]. In fact, other research groups have also carried out the His-to-Gln conversion to show the role of His in the HGSD motif in the biochemical activities of the NDK of Mycobacterium tuberculosis (MtuNDK) [20] and of Bacillus anthracis (BanNDK) [47]. In view of these reasons, residues, such as Ala or Gly, were not considered as alternative to Gln for mutagenesis.
The NTPase, NTP synthesising, and autophosphorylation activities of MsmNDK are similar to those of NDKs from other bacterial genera and other organisms [20, 25, 30, 32, 48]. Although, the NTP synthesising activity of MsmNDK had earlier been shown [18], the requirement of H117 for these activities had not been demonstrated. The lack of complete abolition of GTPase activity in MsmNDK-H117Q might be indicative of other residues being involved in GTP hydrolysis. Studies using NDK mutants of M. tuberculosis (MtuNDK) indicated that besides His117, the residues, Lys10, His53, Tyr50, Arg86, and Arg104, are also involved in either phosphotransfer or autophosphorylation activity [28]. Incidentally, these five residues are conserved between MsmNDK and MtuNDK. This shows the strong possibility of the involvement of the additional residues in the biochemical activities, such as in the GTPase activity, of MsmNDK. Although MsmNDK binds GTP and hydrolyses it, the protein lacks the GXXGK and DXXG motifs of GTP binding proteins [49, 50], and NKKD, which is known to be involved in guanine base recognition [51], like in the case of MtuNDK [20].
Besides the histidine autophosphorylation, the in vivo serine phosphorylation of human NDK, NM23-H1, was found to correlate with the suppression of tumour metastatic potential in the TK melanoma cells, suggesting its biological relevance [52]. Similar serine residue phosphorylation was observed in the NDKs of rat mucosal mast cells [33], Myxococcus xanthus [30], Dictyostelium discoideum [53], and Solanum chacoense [54]. Except in Myxococcus xanthus [30], autophosphorylation of non-histidine residues has not been reported for NDKs from bacterial systems. Serine autophosphorylation of MsmNDK was not found in vitro. Taken together, the observations in the present study show that MsmNDK possesses all the biochemical activities that are characteristic of NDKs from other bacterial genera and that H117 in the HGSD motif is required for the biochemical activities.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGEMENTS
This work was supported by a Centre of Excellence in Tuberculosis Research part-grant from DBT. MA was a CSIR SRF. The infrastructural facilities provided by the DBT Programme Support – Pathogen Biology - to Biological Sciences Division, IISc, UGC-CAS and DST-FIST support to the MCB Dep’t., IISc, are acknowledged.
REFERENCES
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]


