Effects of α-Particle Radiation on MicroRNA Responses in Human Cell-Lines
Abstract
A variety of alpha (α)-particle emitters are found ubiquitously in the environment, in commercial/therapeutic prod-ucts and are a potential threat in the form of a radiological dispersal device. Our understanding of the biological mechanisms and long-term health effects resulting from α-particle exposure is limited. Exposure to radiation induces modulations of gene networks, possibly through microRNAs (miRNAs), which could be targets for studying biological effects. In this study, changes in miRNA expression patterns after 0.5 Gy, 1.0 Gy and 1.5 Gy of α-particle radiation at a low dose-rate of exposure in three human cell-lines (A549, THP-1 and HFL) were investigated. The screening of 1,145 miRNAs across three human cell-lines resulted in unique, cell-specific responses with no overlap in miRNA expression observed in the three cell-lines. Prediction analysis suggests these α-particle induced miRNA mapped to target genes related to ribosomal assembly, lung carcinoma development, cell communication and keratin sulfate biosynthesis. Taken together, these results suggest that exposure to α-particle radiation results in cell-type specific responses in gene network regulatory processes.
INTRODUCTION
In recent years, α-particle radiation has become a rising public health concern. Despite their limited penetrating power, α-particles have a strong capacity to produce an intensely damaging biological response due to their dense ionization tracks that can directly break chemical bonds [1-2]. For this reason, α-particle ingestion (e.g. Polonium-210) or inhalation (e.g. radon (Rn-222) gas, cigarette smoke) may have detrimental effects leading to potential long-term health consequences. Furthermore, α-particle emitters like Americium-241, Plutonium-238 and Polonium-210 have been identified by the American Nuclear Regulatory Commission and the Secretary of Energy (www.energy.gov/media/RDDRPTF14MAYa.pdf) as the most probable isotopes to be used in radiological dispersal devices (RDD). Although numerous epidemiological studies have shown considerable evidence associating exposure to α-particle radiation with adverse health effects (e.g. lung cancer) [3-6], clear mechanistic pathways leading to these effects have yet to be elucidated.
Currently, there is a large amount of data regarding the cytogenetic effects of α-particle radiation [7], only a few selected studies have examined the transcriptional/proteomic effects and no studies to date have examined microRNA (miRNA) expression. miRNAs are small RNAs of approximately 22 nucleotides in length that regulate the expression of complementary messenger RNAs by base pairing with target mRNAs. miRNAs account for 1% of predicted genes in higher eukaryotic genomes and only very few miRNAs have been functionally delineated [8]. These have been shown to have varying roles including involvement in haematopoiesis, cell death, cell proliferation, and in the development of certain cancer types [9]. Furthermore, it has also been shown using in vitro and animal based studies that miRNAs are the regulators of cellular responses to ionizing radiation.
In 2009, Shin et al., [10] showed that epithelial cells exposed to 20 Gy and 40 Gy of γ-radiation (Cesium (Cs)-137 source) caused alterations in 12 and 18 miRNA respectively. These were found to be involved in apoptosis, cell cycle regulation and DNA damage. In a recent investigation where mouse fibroblasts were left for 2 h after exposure to 7.5 Gy of Cs-137 radiation, a decrease in miRNA levels was observed in comparison to embryonic stem cells which showed increased miRNA expression following irradiation [11]. In contrast, miRNA expression profile analysis in transformed lymphoblasts exposed to 2.5 Gy of γ-irradiation displayed minor effects at the miRNA level 4 h post-exposure [12]. The culmination of these studies suggests that radiation is inducing biological effects on gene networks, however the responses vary with cell type, dose and radiation type. A majority of these studies have focused efforts on γ- and X-irradiation. Compared with γ- or X-ray (low linear energy transfer (LET) radiation), high-LET radiation-induced damage (generated by α-particles) is more difficult to repair [13]. With the prevalence of α-particles in the form of Rn-222 gas and their current use in cancer treatment therapies, understanding long-term biological effects are of importance. No studies to date have assessed the biological impacts induced by α-particle radiation at the level of miRNA regulated gene networks.
The aim of the present study was to identify miRNAs responding to low to moderate doses of α-particle radiation in a dose- dependent manner as they would represent reliable radiation responsive targets. These responding miRNAs would provide an insight into regulatory networks induced by α-particle exposure. For this purpose, three human-derived cell-lines were exposed to Am-241 electroplated discs, at doses 0.5 Gy, 0.1 Gy and 1.5 Gy and harvested 24 h post-exposure and analyzed for differential changes in miRNA expression patterns relative to a control group.
MATERIALS AND METHODS
Cell Exposure and Harvesting
A human lung epithelial cell line (A549), a peripheral monocytic cell line (THP-1) and human primary lung fibroblasts (HFL-1) were obtained from The American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in a humidified incubator (37oC, 5% CO2/95% air) in 75 cm2 tissue culture flasks (Costar, Cambridge, MA, USA). THP-1 cells were grown to 90% confluence for 2-3 days in Royal Park Medical Institute (RPMI)-1640 (Invitrogen Canada, Burlington, ON Canada) in media containing 10% fetal bovine serum (FBS) (Sigma-Aldrich Canada, Oakville, ON, Canada). HFL-1 and A549 cells were cultivated in F-12K medium (Invitrogen), containing 10% (FBS) (Sigma-Aldrich Canada). The cells were cultured to ~ 90% confluency, then exposed to α-particle radiation at doses ranging from 0.0 (control) to 1.5 Gy, using Am-241 electroplated discs(Eckert and Ziegler Isotope Products Ltd, Valencia, CA, USA) having an activity level of 66.0 kBq ± 3% (dose rate of 0.98 ± 0.01 Gy/h, LET of 127.4±0.4 keV/µm). The absorbed dose of α-radiation to which cells were exposed was calculated using the GEANT4 v.9.1 Monte Carlo toolkit and validated with in vitro experiments [14]. For the α-particle exposures, cells were cultured in thin Mylar based plastic dishes (MD) (Chemplex Industries, Palm City, FL, USA), which allowed the penetration of the α-particles. Cell viability was assessed from a 30 µL aliquot by the dual stain viability assay [15] both prior to exposure, 24 h post-exposure and a further 72 h post-exposure for THP-1 cells at all doses tested. The remaining cells were centrifuged at 1000 rpm for 5 min, decanted, and resuspended in 350 µL of lysis buffer, provided by Qiagen’s RNeasy Mini kit. The cells were stored at -80oC until RNA isolation (Qiagen Inc, Mississauga, ON, Canada).
RNA Isolation
The frozen lysates were pipetted onto a QIAshredder spin column, and the miRNA was extracted using the miRNeasy Mini kit according to the manufacturer’s instructions (Qiagen Inc.). The concentration and quality of the RNA sample isolation was determined through spectrophotometric means (optical density (OD) ratio of A260:A280), using the Agilent 2100 Bioanalyzer, following the manufacturer’s instructions (Agilent Technologies, Mississauga, ON). Samples with a RIN value of greater than or equal to 8.0 were deemed to be acceptable for analysis. An input of 200 ng of RNA was used for miRNA analysis following the Illumina(r) MicroRNA Expression Profiling Assay Guide (11317302 Rev. A). Samples were hybridized on Illumina human v2 miRNA BeadChips. BeadChips were imaged and quantified with the Illumina iScan scanner and data was processed with Illumina GenomeStudio v2010.2.
Statistical Analysis
Data pre-processing was done within GenomeStudio, where the intensities were averaged per probe/miRNA. Normalization of dataset was conducted in GeneSpring (version GX 11.5). Intensities were normalized to the 25th percentile. Intensities were log2 transformed and a two tailed T-test was performed. The variance was not assumed to be the same between the groups. Multiple testing using Benjamini & Hochberg false discovery correction was applied to the p-values in order to obtain robust responding miRNA targets.
Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
Selected genes identified by miRNA array analysis as displaying statistical significance, with fold changes of 2 or higher and for which primers were validated were further assessed by qRT-PCR. Total RNA (100 ng) isolated from cells was reverse transcribed into complementary DNA using the RT2 First Strand Kit (SABiosciences Corp., Frederick, Maryland, USA). Gene profiling was performed according to the manufacturer’s instructions using custom RT2-profiler PCR arrays (SABioSciences Corp.). Reactions were prepared in 96-well plates and performed in duplicate in a spectrofluorometric thermal cycler (Biorad iCycler; Hercules, CA). The relative expression of each gene was determined by using the comparative threshold (Ct) method [16]. Analysis of qRT-PCR expression profiles and statistical analysis of data were performed using the super array biosciences web portal for data analysis of their products. (SABiosciences http://www.sabiosciences.com/pcr/arrayanalysis.php).
Target Prediction Analysis
Predicted transcript targets of significantly altered miRNA (p≤0.05, FC≥|1.5| for all doses, Table 1) were found using Microcosm v5 (maintained by the European Bioinformatics Institute, http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/tar-gets/v5/). These unique transcript identifiers (ENST) were then matched to their corresponding genes by performing a biomart data query (maintained by Ontario Institute for Cancer Research and the European Bioinformatics Institute, http://www. biomart.org/). Unique gene identifiers (ENSG) were assigned using the “ensemble genes v62” and the “homo sapiens genes GRCh37.p3” search parameters. The ENST numbers were input as an ID select filter with output attributes of ENSG and ENST. These ENSG gene lists were then submitted as gene candidates to the DIANA-mirPath algorithm to find specific pathways that may be modified by miRNA modulations. The queries were submitted to the DIANA-mirPath multiple miRNA analysis using the DIANA MicroT v4 search algorithm [17] as assembled gene lists from each respective cell type. Pathways are identified by contrasting the genes submitted compared to the genes present in the Kyoto Encyclopaedia of Genes and Genomes KEGG canonical pathways. Only significant (p≤0.05) pathways generated from the union of cell-specific gene-lists are reported.
miRNAs responsive at all doses from array profiling. Statistically significant differentially expressed miRNA with corresponding p-values and fold changes (FC) which were found to respond at all doses of α-particle radiation tested (0.5, 1.0, and 1.5 Gy) and were harvested 24 hours post-exposure. Statistical cut-off at p ≤ .05, FC≥|1.5| with n = 5 biological replicates
| Cell-Type | miRNA | 0.5 Gy FC | p-Value | 1.0 Gy FC | p-Value | 1.5 Gy FC | p-Value |
|---|---|---|---|---|---|---|---|
| A549 | hsa-miR-486-5p | 1.94 | 0.00 | 2.57 | 0.00 | 2.91 | 0.00 |
| hsa-miR-187 | 1.54 | 0.00 | 1.81 | 0.00 | 2.00 | 0.00 | |
| hsa-miR-25* | -1.69 | 0.02 | -1.69 | 0.02 | -1.67 | 0.02 | |
| hsa-miR-92a-1* | -1.84 | 0.00 | -1.93 | 0.00 | -2.11 | 0.00 | |
| hsa-miR-424* | -2.21 | 0.00 | -1.79 | 0.01 | -1.87 | 0.00 | |
| hsa-miR-16-1* | -1.61 | 0.01 | -1.56 | 0.01 | -1.63 | 0.00 | |
| hsa-miR-23a* | -1.73 | 0.01 | -1.54 | 0.03 | -1.66 | 0.01 | |
| HFL-1 | hsa-miR-663 | 2.87 | 0.01 | 2.42 | 0.02 | 2.19 | 0.01 |
| hsa-miR-491-5p | 1.47 | 0.03 | 1.64 | 0.01 | 1.61 | 0.01 | |
| hsa-miR-560:9.1 | 2.47 | 0.00 | 2.48 | 0.00 | 2.17 | 0.01 | |
| hsa-miR-675 | 2.75 | 0.00 | 2.34 | 0.05 | 1.79 | 0.05 | |
| hsa-miR-1228* | 1.53 | 0.01 | 1.54 | 0.01 | 1.58 | 0.01 | |
| THP-1 | hsa-miR-708 | 2.03 | 0.02 | 2.16 | 0.02 | 2.51 | 0.00 |
* an miRNA expressed at low levels relative to the miRNA in the opposite arm of a hairpin
RESULTS
Cellular Viability
The viability and cell numbers were determined 24 h post-exposure and a further 72 h post-exposure for THP-1 cells. No significant effects on viability or cell number were observed at any doses tested in this study. The viability remained above 98% in all treatment groups and controls, indicating that the α-particle radiation did not cause a significant degree of cell death (data not shown). Previous studies from our laboratory have assessed the biological impacts of α-particle radiation effects on cellular viability, apoptosis, cell survival and γ-H2AX induction [14, 18]. Immediate effects on cell viability are minimal; however DNA damage induction can be seen 30 minutes post-exposure through γ-H2AX assessment [19]. Longer-term viability and cell survival effects were apparent when measured through apoptotic induction (96h) (caspase activation and annexin V binding) and clonogenic assay (2 weeks), respectively.
miRNA Profiling
miRNA profiling was assessed in three cell-lines following exposure to moderate doses of α-particle radiation. Only highly significant miRNA expressions are presented which were observed across the three doses tested, and with fold change > |1.5|. Among the 1,145 miRNAs that were screened in the three cell-lines, only a selected few were shown to be statistically significant and were found to be uniquely responding by cell-type (Table 1). A549 cells showed the highest number of responding miRNAs, a total of 7 miRNAs were expressed at the three doses tested, 71% were downregulated and 29% were shown to be upregulated. Highest expression was observed with miR-486-5p and the lowest expression was observed with miR-424*. Human lung fibroblasts exposed to α-particle radiation expressed 5 responding miRNAs, all of which were upregulated. Among the 5 miRNAs, three (miR-663, miR-560:9.1 and miR-675) were shown to consistently have 2 fold or higher levels of expression. Screening of THP-1 cells following radiation insult resulted in no statistically significant responses 24h post-exposure. However, the induction of miR-708 was observed three days post-exposure. A heat map was constructed to provide a qualitative representation of the similarities and differences in expression patterns of the miR- responses obtained for the different cell-types (Fig. 1). The heat map further highlights the unique responses obtained for each cell-type. A few miRNAs were expressed consistently at all three doses (miR-25*, -16-1*, -491-5p, -1228*), while other miRNAs exhibited a graded dose-responsive expression (miR-486-5p, -9-29-1*, -424*, -663, -675, -708).
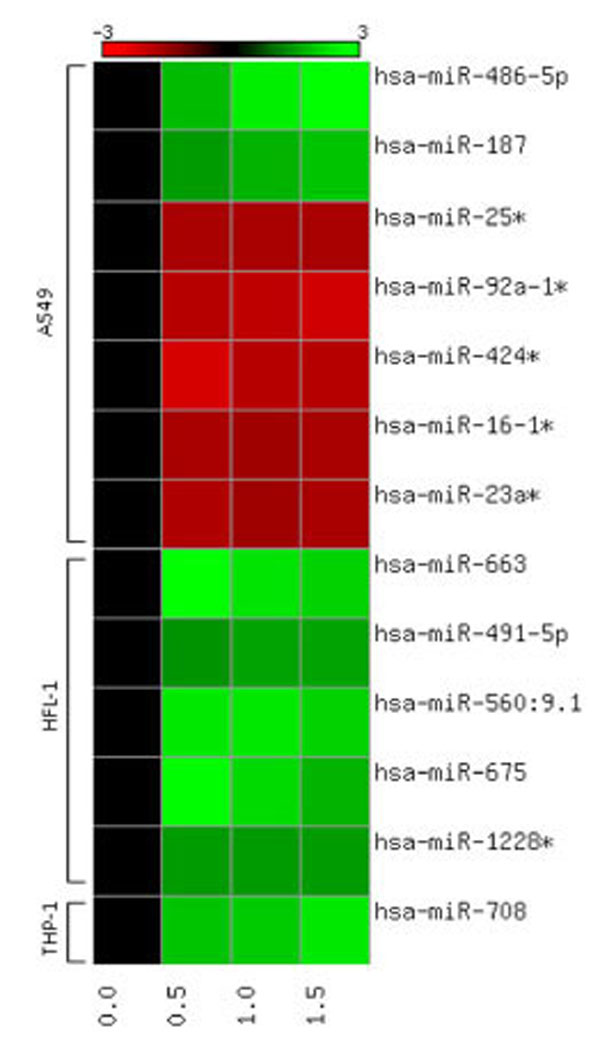
Heat map showing quantitative expression patterns of miRNAs responsive at all doses (0.0, 0.5, 1.0, 1.5 Gy) for all cell lines (A549, HFL-1 THP-1). Red signifies down-regulation and green signifies up-regulation in miRNA expression, based on an n=5 biological replicates.
Validation of Altered Expression of Radiation-Inducible miRNA
A few select dose-responsive miRNAs were further validated using qRT-PCR. As shown in Table 2, a comparable pattern of expression and significance was observed across all doses tested using the two methodologies. Therefore, the miRNA responses are robust and valid targets for further analysis studies.
qRT-PCR validation of array response. qRT-PCR was conducted using custom designed primer plates to validate the response of the miRNA arrays. Statistically significant differentially expressed miRNA with corresponding p-values and fold changes (FC) which were found to respond at all doses of α-particle radiation tested (0.5, 1.0, and 1.5Gy). Statistical cut-off at p ≤ 0.05
| Cell-Type | µRNA | 0.5 Gy FC | SEM | 1.0 Gy FC | SEM | 1.5 Gy FC | SEM |
|---|---|---|---|---|---|---|---|
| A549 | hsa-miR-486-5p | 2.02 | 0.30 | 1.86 | 0.15 | 2.21 | 0.21 |
| HFL-1 | hsa-miR-675 | 2.06 | 0.25 | 2.10 | 0.15 | 2.30 | 0.22 |
| hsa-miR-663 | 2.10 | 0.26 | 2.01 | 0.14 | 2.14 | 0.31 | |
| hsa-miR-560:9.1 | 2.15 | 0.31 | 2.08 | 0.19 | 2.2 | 0.14 | |
| THP-1 | hsa-miR-708 | 2.00 | 0.26 | 2.20 | 0.16 | 2.60 | 0.16 |
Target Prediction Analysis
DIANA-mirPath is a web-based computational tool that was used to identify molecular pathways potentially altered by the expression of the multiple miRNAs and their associated genes that were obtained for each cell-line. The analysis tool allowed miRNA target genes to be compared to a set of miRNA targets to all known KEGG pathways. The combinatorial effect of co-expressed microRNAs for each cell-line showed uniquely responding pathways to be activated. Submission of significantly modulated miRNA gene lists to the DIANA-mirPath algorithm resulted in the identification of statistically significant KEGG pathways (Table 3). Target prediction analysis of miRNA from epithelial lung cell (A549) identified a total of four statistically significant canonical pathways including ribosome assembly, small cell lung cancer formation and the transforming growth factor (TGF)-β signaling cascade. Submission of gene lists from the human lung fibroblasts (HFL-1) identified nine pathways, the most statistically significant being cell communication. Human monocytic cell (THP-1) analysis yielded three significant pathways, including those involved in keratin sulfate biosynthesis, hematopoietic cell lineage and peroxisome proliferator-activated receptor (PPAR) signaling. The ribosome assembly pathway was the only one that was shown to be common to both A549 and HFL-1 cells.
Statistically significant pathways as identified by the DIANA-mirPath multiple miRNA algorithm. Pathways were found using gene targets of responding miRNAs
| Cell-Type | Pathway | KEGG Pathway Code | # Genes Gene | Targets | p-Value |
|---|---|---|---|---|---|
| A549 | Ribosome | hsa03010 | 4 | RPL35 RPL3L RPS5 RPS6 | 0.002 |
| Small cell lung cancer | hsa05222 | 8 | AKT3 CCNE1 LAMA5 AMB3 NFKB2 PTK2 RELA RXRB | 0.028 | |
| gamma-Hexachlorocyclohexane degradation | hsa00361 | 9 | ACP5 ACP6 ACPP ALPI ALPL ALPPL2 DHRS2 PON1 PON2 | 0.035 | |
| TGF-beta signaling pathway | hsa04350 | 9 | ACVR2B COMP ID3 MAPK3 NOG TGFB1 THBS4 TNF ZFYVE9 | 0.037 | |
| HFL-1 | Cell Communication | hsa01430 | 24 | CHAD COL11A2 COL1A1 COL6A1 COL6A2 COMP ITGB4 KRT14 KRT15 KRT17 KRT23 KRT37 KRT39 KRT5 KRT71 KRT76 KRT82 KRT9 LAMA1 LAMA5 LAMB3 LAMB4 NES TNC | 0.003 |
| Phosphatidylinositol signaling system | hsa04070 | 14 | CALML3 CDIPT DGKA INPP5E ITPKA ITPR3 PI4KB PIB5PA PIK3CA PIK3CD PIP5K1B PLCG2 PLCZ1 PRKCG | 0.011 | |
| Lysine degradation | hsa00310 | 9 | DLST ECHS1 EHMT2 HADHA NSD1 OGDH OGDHL PLOD1 PLOD3 | 0.013 | |
| Inositol phosphate metabolism | hsa00562 | 10 | INPP5E ITPKA MIOX PI4KB PIB5PA PIK3CA PIK3CD PIP5K1B PLCG2 PLCZ1 | 0.022 | |
| Glycan structures - biosynthesis 1 | hsa01030 | 4 | ALG12 B4GALT3 CHPF ST3GAL2 | 0.023 | |
| Focal adhesion | hsa04510 | 28 | ACTN1 BAD BCAR1 CHAD COL11A2 COL1A1 COL6A1 COL6A2 COMP FLNA ILK ITGA2B ITGB4 LAMA1 LAMA5 LAMB3 LAMB4 MYL2 MYL9 MYLK2 MYLPF PIK3CA PIK3CD PPP1CA PRKCG RAC3 TLN1 TNC | 0.031 | |
| Leukocyte transendothelial mi-gration | hsa04670 | 18 | ACTN1 BCAR1 CD99 CLDN15 CLDN4 CYBA MAPK12 MAPK13 MMP9 MYL2 MYL9 MYLPF NCF1 PIK3CA PIK3CD PLCG2 PRKCG RAPGEF3 | 0.033 | |
| Ribosome | hsa03010 | 2 | RPL3L RPS11 | 0.035 | |
| ECM-receptor interaction | hsa04512 | 14 | AGRN CHAD COL11A2 COL1A1 COL6A1 COL6A2 ITGA2B ITGB4 LAMA1 LAMA5 LAMB3 LAMB4 SV2A TNC | 0.041 | |
| THP-1 | Keratan sulfate biosynthesis | hsa00533 | 4 | ST3GAL3 FUT8 B4GALT2 B4GALT3 | 0.000 |
| Hematopoietic cell lineage | hsa04640 | 7 | CD33 CD1A CD38 CD34 IL4R ITGA5 HLA-DRB5 | 0.024 | |
| PPAR signaling pathway | hsa03320 | 6 | APOC3 RXRB APOA5 APOA2 FABP3 ACOX3 | 0.039 |
DISCUSSION
MiRNAs are involved in many biological processes and dysregulation of these networks can result in diseases such as cancer. It has been known for decades that ionizing radiation promotes carcinogenesis and the impact of a high-linear energy transfer (LET) radiation type such as α-particle is more potent in comparison to X-ray and γ-irradiation [20,21]. Due to these damaging qualities, α-particles have emerged as promising radiation types for use in cancer treatment therapies [22] and more undesirably as isotopes to be used in RDDs. Therefore, understanding their biological impact is important. At the cellular level, it is well known that α-particle radiation causes DNA double strand breaks which are more difficult for the body to repair [13]. An understanding of this response at the gene network regulatory level is limited. Therefore, the focus of this study was to assess in three cell-lines the effects of α-particle radiation on miRNA expression. Specifically, two human lung cell-lines were assessed as these may be relevant to environmental/inhalation exposures, i.e., through Rn-222 gas inhalation. In addition, a blood monocytic cell-line was examined which may be more pertinent for biomarker discovery.
Overall, alterations in miRNA profiles caused by α-particle radiation resulted in a limited response in all three cell-lines that were assessed. Of the responding miRNAs, all were shown to be responsive at the three doses tested. A549 cells exposed to 0.5-1.5 Gy of radiation displayed the expression of 7 miRNAs. A majority of these were shown to be downregulated. MiR-486-5p was significantly upregulated by ~3 fold at the high dose tested. miR- 486-5p has been consistently shown to be differentially expressed in various cancer types [23]. Among the downregulated miRNAs miR-92a-1 emerged as being decreased in expression by 2 fold at all three doses tested. Overall, target prediction of the differentially expressed miRNAs obtained for A549 cells showed them to be involved in four pathways including ribosome assembly, small cell lung cancer, gamma-hexacheorocyclohexane degradation and TGF-beta signaling. A study in 2009, irradiating A549 cells with γ-rays from a Cs-137 source showed the expression of 20 miRNAs with fold changes greater then 2 fold [10]. However, irradiations were conducted at 20 and 40 Gy, which is significantly higher from that which was used in this study. In fact, no overlapping miRNA responses were seen between the Shin et al. [10] study and this investigation. This further highlights that a low-LET radiation type may activate different responding pathways in comparison to high-LET radiation exposure as observed for α-particle radiation.
MiRNA analysis of human lung fibroblasts showed the expression of 5 α-particle induced-miRNAs, all of which were shown to be upregulated. miR-663 displayed the highest fold change relative to the other induced miRNAs, though interestingly, the expression was shown to decrease with the dose of radiation. This was most noticeable with miR-675, which showed the highest level of expression at the lowest dose of radiation. This would imply that higher doses of radiation may cause degradation of miRNA products in order to allow for gene expression. Target prediction analysis showed that most statistically significant pathways associated with miR-675 are in cell communication; specific gene targets using Microcosm analysis include COMP, ITGB4, KRT23, LAMA1, NES and TNC. A study by Maes et al. on [24] profiled normal human skin fibroblasts for miRNA expression changes in a time course after 0.1 Gy and 2.0 Gy of X-rays. Contrasting their results with this investigation showed only one miRNA, miR-663, to be common between the two studies. However, in the Meas et al. investigation, this miRNA was shown to be downregulated after 2.0 Gy of X ray and 0.5 h post-exposure whereas in this study a marked increase in miRNA was observed following irradiation with 0.5-1.5 Gy of α-particle radiation. The lack of commonality and reciprocal regulation of the same miRNA suggests that a different regulatory response is elicited by the two radiation types, despite the exposure to a similar dose of radiation.
To determine if the responses observed for HFL-1 and A459 cells were induced in a circulating cell-type, which is more relevant to the identification of a biodosimeteric marker for α-particle radiation, THP-1 cells were assessed for miRNA responses following irradiation at equivalent doses. Interestingly, no statistically significant miRNA effects were observed 24 h post-exposure. This was not unexpected as previous studies have shown that THP-1 cells produce delayed responses in gene expression following exposure to an insult [18]. A study conducted in our laboratory has shown that THP-1 cells exposed to α-particle radiation at 0, 0.5, 1.0, and 1.5 Gy of radiation show no effects on apoptosis and gene expression 24 h post-exposure [18]. Significant changes were only observed 96 h post-exposure. Therefore, in this study, further analysis at 72 h showed expression of 1 statistically significant miRNA. A strong 2 fold dose-responsive increase in the expression of miR-708 was observed, which was further validated by qRT-PCR. Interestingly, miR-708 has emerged as an important early detection marker of squamous cell lung cancer in sputum [25]. Using DIANA analysis, miR-708 gene targets were predicted to be involved in three statistically significant pathways including keratin sulfate biosynthesis, hematopoietic cell-lineage and PPAR signaling pathway. Keratin sulfate pathway was shown to be the top statistically significant canonical pathway that may be targeted by miR-708. This pathway has been shown to involve genes ST3GAL3, FUT8, B4ALT2 and B4GALT3 and is a component of the glycan metabolism and biosynthesis pathway. As miR-708 was uniquely induced, it may lend itself to be a candidate biomarker of α-particle radiation response. However, further analysis and verification of this response would be needed using lymphocytes from healthy individuals following α-particle exposure.
In conclusion, this study highlights a subset of human miRNAs that show robust expression changes in response to α-particle radiation. To date, no studies have examined the responses of three cell-types on miRNA expression following α-particle radiation. All cell-types that were assessed in this study showed differentially responding gene regulators, the lung epithelial cell-line displaying the greatest number of responding miRNAs. In addition, we have potentially identified a bio-dosimeteric marker, miR-708, which was only expressed in a circulating blood monocyte and exhibited a delayed induction. This marker may be a basis for further validation studies. Overall, this study has identified α-particle induced miRNAs that may be associated with adverse biological effects related to carcinogenesis; however, it should be noted that any possible effects of α-particle radiation on miRNA expression cannot, as of this time, be directly correlated to human disease. Further validation would be required to identify the specific pathways that are involved in these regulatory responses.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENTS
This work was supported by the Health Canada Research and Development Genomic Initiative. The authors are grateful to the Vancouver Prostate Centre for processing of the miRNA arrays.
ABBREVIATIONS
REFERENCES
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]


