All published articles of this journal are available on ScienceDirect.
Transcriptional and Secretomic Profiling of Epidermal Cells Exposed to Alpha Particle Radiation
Abstract
Alpha (α)-particle emitters are probable isotopes to be used in a terrorist attack. The development of biological assessment tools to identify those who have handled these difficult to detect materials would be an asset to our current forensic capacity. In this study, for the purposes of biomarker discovery, human keratinocytes were exposed to α-particle and X-radiation (0.98 Gy/h at 0, 0.5, 1.0, 1.5 Gy) and assessed for differential gene and protein expression using microarray and Bio-Plex technology, respectively. Secretomic analysis of supernatants showed expression of two pro-inflammatory cytokines (IL-13 and PDGF-bb) to be exclusively affected in α-particle exposed cells. The highest dose of α-particle radiation modulated a total of 67 transcripts (fold change>|1.5|, (False discovery rate) FDR<0.05) in exposed cells. Several genes which responded with high expression levels (>2 fold) included KIF20A, NEFM, C7orf10, HIST1H2BD, BMP6, and HIST1H2AC. Among the high expressing genes, five (CCNB2, BUB1, NEK2, CDC20, AURKA) were also differentially expressed at the medium (1.0 Gy) dose however, these genes were unmodulated following exposure to X-irradiation. Networks of these genes clustered around tumor protein-53 and transforming growth factor-beta signaling. This study has identified some potential gene /protein responses and networks that may be validated further to confirm their specificity and potential to be signature biomarkers of α-particle exposure.
INTRODUCTION
Increased forensic capability through the development of biological tools to help identify those involved in a terrorist precipitated attack should be an integral part of the strategy against terrorism. Such capabilities have the potential to deter a mass casualty event by possibly incriminating those involved prior to the incident. In addition, these capacities would be of benefit for rapidly identifying those who are exposed, if such a terrorist event were to occur (American Physical Society http://www.aps.org/policy/reports/upload/Nuclear-Forensics-Report-FINAL.pdf). Over the decades, the use of gene-based markers in the development of forensic tools has emerged as an attractive alternative to classical cytogenetic assays [1]. Radiation insults have been shown to result in modulations in the expression of genes [2] proteins/cytokines [3] and microRNA [4]. Furthermore, different radiation types can elicit exclusive effects that could represent signature responses to a specific radiation type [5].
Currently the development of gene based bio-chips to low linear energy transfer (LET) radiation (gamma, x-ray) is un- derway [6]. There is potential to successfully discriminate be tween exposed and non-exposed individuals using gene expression profiling from a fingerpick of blood (Patent # WO2000050643). However, similar availability of tools for high LET radiation-types (alpha, neutrons, heavy ions) remains limited. Alpha particle emitting isotopes such as Americium-241 have been identified as being amongst the most probable to be used in a radiological dispersal device [7]. In such a scenario, external dermal contamination would be a most likely exposure route to the general public. Settled α-particles can be highly damaging to sensitive exposed skin cells, such as on the face, neck or hands [8]. Furthermore, dose estimates have been constructed for various settled α-particle emitters on the skin and the dose to the basal layers of the skin have been estimated at 0.5-1µSv/decay/cm2 with an energy deposition of about 0.5 Gy per α- particle track through the nucleus [9].
Keratinocytes are the predominant cell type in the epidermis, the outermost layer of the skin, constituting ~95% of the cell population [10]. They can elicit a series of inflammatory response cascades that can affect the gene and protein microenvironment. Keratinocytes have been shown to respond to radiation insults such as UV radiation with the production of pro-inflammatory mediators and gene responses [reviewed in 11, 12]. Therefore they serve as a pertinent cell-type to monitor effects associated with α-particle radiation exposures. To date, there have been no studies examining genomic or secretomic screening of keratinocytes following α-particle radiation insult. A selected few studies have shown effects on specific gene markers following the exposure of various tissue cells to α-particle radiation using in vitro and animal based models [13, and 14]. Previous work in our laboratory has identified a series of gene-based markers that are modulated by exposure of human lung cells and blood cells to α-particles [15]. In terms of protein analysis, interleukin-8 (IL-8) has been shown to be affected by high-LET radiation [16]. Intracellular protein modulations were also observed in the lungs of rats exposed to radon gas (an α-particle emitter) [17]. Previous studies in our lab have shown the modulation of pro-inflammatory cytokines in α-particle exposed lung derived cells and not in X-irradiated blood suggesting the potential for differential expression among radiation types [18].
The aim of the present study was to identify gene/protein based biomarkers responding to low to moderate doses of α-particle radiation in a dose-dependent manner within the over-arching goal of developing a forensic tool to identify individuals that have handled special nuclear materials or have had dermal exposure to α-particle emitting isotopes. For this purpose, human-derived keratinocytes were exposed to α-particles and analyzed for differential changes in gene expression patterns and the secretion of a series of pro-inflammatory cytokines and growth factors.
MATERIALS AND METHODS
Cell Exposure and Harvesting
Primary human derived keratinocytes (HEKn) obtained from the Invitrogen (ATCC, Manassas, VA, USA), were maintained in a humidified incubator at 37°C, 5% CO2, 95% air in 75 cm2 flasks (T-75). The cells were grown in EpiLife media (Invitrogen) containing Human Epidermal Growth Supplements (Invitrogen) as per manufacturer’s instructions. Cells were seeded onto 35 mm culture dishes containing 2 layers of 2.5 μm-thick mylar films at a density of 2.5 x 105 cells with 5 ml of culture media and allowed to grow to 90% confluency over 3 days. Cells were then washed using Phosphate Buffered Saline (PBS) and an additional 5 ml of fresh media was added prior to α-particle radiation exposure. These exposures were performed at doses of 0 (control), 0.5, 1.0 or 1.5 Gy using americium (241Am) electroplated discs with an activity level of 66.0 kBq ± 3% (dose rate of 0.98 ± 0.01 Gy/h, LET of 127.4±0.4 keV/µm). The absorbed dose of α-particle radiation to which the cells were exposed was calculated using the GEANT4 v.9.1 Monte Carlo toolkit [19]. For the α-particle exposures, cells were cultured in thin mylar based plastic dishes (MD) (Chemplex Industries, Palm City, FL, USA), which allowed for penetration of α-particles. Cells destined for X-radiation at doses of 0 Gy to 1.5 Gy were exposed using the X-RAD 320 X-ray irradiation system at a low dose rate of 0.98 ± 0.05 Gy/h, 120 keV (Precision X-ray, Inc., North Branford, CT, USA ). Following irradiation, cells were either washed and harvested 24 h after exposure in 350 μl RLT buffer (Qiagen Inc, Mississauga, ON), for transcriptional profiling or 200 μl of media was repeatedly sampled at 24, 48, 72 and 96 h for secretomic analysis. All samples were flash frozen and stored at -80°C until ready for use. For each exposure group and end point, a total of 5 independent experiments were conducted. Cell viability was assessed 96 h post-exposure via Trypan Blue (Bio-Rad) viability assay. After 96 h both the control and 1.5 Gy α- and X-ray exposed cells remained ~90% viable (data not shown).
Microarrays
Frozen cell lysates were pipetted onto a QIAshredder spin column, and total RNA was extracted using the RNeasy Mini kit according to the manufacturer’s instructions (Qiagen Inc.). Additionally, Qiagen’s On-Column RNase-free DNase was used to eliminate possible DNA contamination. The concentration and quality of the RNA sample isolation was determined through spectrophotometric means (optical density (OD) ratio of A260:A280), and using an Agilent 2100 Bioanalyzer, following the manufacturer’s instructions. All RNA Integrity Numbers (RINs) were above 9.0. An input of 200 ng of total RNA was used for whole genome analysis following the Illumina(r) Whole Genome Expression Profiling Assay Guide (11317302 Rev. A). Samples were hybridized on Illumina human-12 v4 RNA BeadChips. BeadChips were imaged and quantified with the Illumina iScan scanner and data were processed with Illumina GenomeStudio v2010.2 software.
The preprocessing includes three steps: background correction (performed in GenomeStudio software: www.illumina. com), quantile normalization [20] and log2 transformation of normalized data. The last two steps were performed in lumi R package [21]. LIMMA (linear models for microarray data) [22, 23] was used to identify differentially expressed gene signatures under different conditions. Briefly, it starts by fitting a linear model for each gene in the data, and then an empirical Bayes (EB) method is used to moderate the standard errors for estimating the moderated t-statistics/F-statistics for each gene, which shrinks the standard errors towards a common value. This test is similar to an ANOVA method for each gene except that the residual standard deviations are moderated across genes to ensure more stable inference for each gene. The moderated standard deviations are a compromise between the individual genewise standard deviations and an overall pooled standard deviation. The false discovery rate (FDR) was evaluated using the Benjamini and Hochberg multiple testing procedure [24].
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
Selected genes identified by microarray analysis as displaying statistical significance, with fold changes of 1.5 or higher and for which primers were validated were further assessed by qRT-PCR. Total RNA (100 ng) isolated from cells were reverse transcribed into complementary DNA using the RT2 First Strand Kit (SABiosciences Corp., Frederick, Maryland, USA). Gene profiling was performed according to the manufacturer instructions using custom RT2-profiler PCR arrays (SABioSciences Corp.). Reactions were prepared in 96-well plates and performed on a spectrofluorometric thermal cycler (Biorad iCycler; Hercules, CA). The relative expression of each gene was determined by using the comparative threshold (Ct) method. Analysis of qRT-PCR expression profiles and statistical analysis of data were performed using the super array biosciences web portal for data analysis of their products.
(SABiosciences http://www.sabiosciences.com/pcr/arrayanaly-sis.php).
Bio-Plex Assay
Supernatants from exposed (0.5, 1.0 and 1.5 Gy α-particle and X-rays) and control samples were analyzed for secretion levels of 27 cytokines starting at 24 h post exposure until 96 hr. Cytokines examined included interleukin (IL)-1b, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, eotaxin, fibroblast growth factor (FGF), Granulocyte-Colony Stimulating Factor (G-CSF), granulocyte/ macrophage colony-stimulating factor (GM-CSF), interferon-γ (IFN-γ), interferon gamma-induced protein 10 kDa (IP-10), monocyte chemotactic protein-1 (MCP-1), macrophage inflammatory protein (MIP)-1a, MIP-1b, platelet derived growth factor (PDGF)-bb, regulated upon activation, normal T-cell expressed, and secreted (RANTES), tumor necrosis factor-α (TNF-α) and vascular endothelial growth factor (VEGF). Analysis was performed using a multiplex suspension array assay according to the manufacturer’s instructions (Bio-Rad). Briefly, conjugated beads were allowed to react with a sample containing a known (standard) or unknown amount of cytokines for 30 min. Conjugated beads with bound target were then washed and incubated with biotinylated detection antibodies that were directed against specific cytokine epitopes. The resulting complexes were then incubated for a further 10 min with streptavidin-phycoerythrin and excess reagent was washed off and assessed for bound cytokine using a microtiter plate reader (Bio-Rad). The concentration of cytokines in supernatants was then assessed from the generated standard curves for each individual cytokine using Bio-Plex software (Bio-Rad). To mine for differentially expressed cytokines the approach of Kleynhans et al. [25] was employed. Briefly, for each radiation type and cytokine, a two-way ANOVA (TIME *DOSE) linear mixed effects model was fit to take the repeated measures into account. Compound symmetry correlation structure was assumed, meaning for all samples the slopes of the lines regressing the dependent variable (intensity of cytokine) were the same over time. DOSE * TIME were treated as a factorial design and used to generate the main effects (DOSE * TIME) and their interaction (DOSE * TIME). Repeated measures at different time-points were treated as random factors.
Pathway Analysis
For the two gene lists (67-gene list and 5-gene list), the significance of predefined pathways, functions and network using Ingenuity Pathway Analysis software (IPA, version 11904312, http://www.ingenuity.com Core Analysis) was conducted. Functional analysis identified the biological functions and/or diseases that were significant to the given gene list. Molecules from the list that were associated with biological functions and/or diseases in the Ingenuity Knowledge Base were considered for the analysis. Right‐tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that list was due to chance alone. Canonical pathway analysis identified the pathways from the IPA library that were significant to the gene list. Molecules from the gene list that were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the gene list and the canonical pathway was measured in two ways: 1) A ratio of the number of molecules from the gene list that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed. 2) Fisher’s exact test was used to calculate a p-value determining the probability that the association between the genes in the list and the canonical pathway is explained by chance alone. The functional analysis of a network identified the biological functions and/or diseases that were significant to the molecules in the network. A network is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity KnowledgeBase. The intensity of the node color indicates the degree of up (red) or down (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Edges are displayed with various labels that describe the nature of the relationship between the nodes (e.g., P for phosphorylation, T for transcription).The network molecules associated with biological functions and/or diseases in the Ingenuity Knowledge Base were considered for the analysis. Similar to the functionality analysis a right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that network was due to chance alone.
RESULTS
Differential Gene Expression Profiles and Pathway Analysis
To mine for differentially expressed genes, LIMMA (22, 23) was employed. In order to reduce the size of the gene lists and to identify more reliable radiation specific genes, significant genes identified by LIMMA (FDR≤0.05) were further filtered on a |1.5| fold change (FC) cut-off. Cells exposed to low dose-rate X-rays at all three doses tested (0.5, 1.0, 1.5 Gy) did not express differentially modulated transcripts that were shown to be statistically significant using Benjamini-Hochberg (BH) corrected p-values relative to the control treatment groups (data not shown). Using less stringent statistical methods, the X-ray treatment did result in changes in gene expression, however these transcripts warrant further validations using alternate methods in order to ensure the responses are robust.
On the contrary, keratinocytes exposed to α-particles showed modulated expression in a number of genes (Table 1). At the lowest dose of α-particle radiation (0.5 Gy) no statistically significant genes were shown to be expressed however the medium dose (1.0 Gy) of radiation caused changes in expression of a total of 5 genes (CDC20, CCNB2, AURKA, BUB1 and NEK2) and all were shown to be upregulated relative to the control treatment group. These 5 genes were also observed to be expressed at the high dose of α-particle radiation (Fig. 1). All 5 of these genes were shown to be upregulated with fold changes approximately in the range of 1.5 fold of the unirradiated control treatment group (Table 2). Pathway analysis of these 5 differentially expressed genes showed that the top high-level functional categories were in relation to DNA replication, recombination and repair, and tissue morphology (FDR≤0.05). Analysis of the low-level functions showed involvement in cell cycle regulation and mitotic repair (Table 3). The top scoring network associated with these genes was centered around TP-53 signaling (Fig. 2).
Number of Transcripts Differentially Modulated (BH p≤ 0.05 FC > |1.5|) at Each Dose of Alpha Particle Radiation and Relative Regulation Patterns
| 0.5 Gy | 1.0 Gy | 1.5 Gy | |
|---|---|---|---|
| Number of Transcripts | 0 | 5 | 68 |
| Common Amongst Doses (%) | N/A | 5(100) | 5(7) |
| Exclusive (%) | N/A | 0 (0) | 63(93) |
| Up Regulated (%) | N/A | 5(100) | 51 (75) |
| Common Amongst Doses (%) | N/A | 5 (100) | 5 (10) |
| Exclusive (%) | N/A | 0 (0) | 46 (90) |
| Down Regulated (%) | N/A | 0(0) | 17(25) |
| Common Amongst All Doses (%) | N/A | 0 (0) | 0 (0) |
| Exclusive (%) | N/A | 0 (0) | 17 (100) |
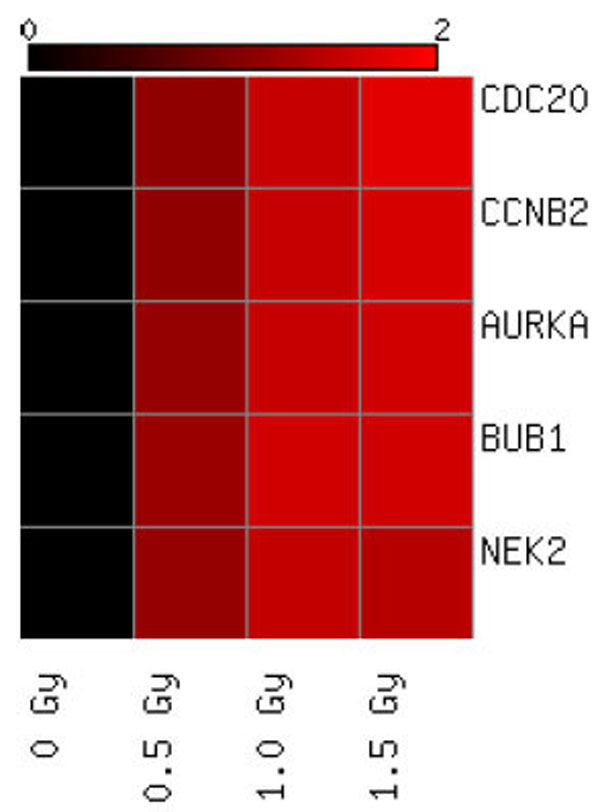
Heat map showing microarray fold change expression values of the 5 genes which were found to be significant at the medium and high doses of radiation exposure using microarray technology. Red colouring signifies up-regulation based on an n=5 biological replicates.
Genes which were Statistically Significant (BH Corrected p Value < 0.05, FC ≥ 1.5) at the 1.0 and 1.5 Gy Dose of α-radiation Sorted by Fold Change at the Highest Dose Level
| 1.0 Gy | 1.5 Gy | |||
|---|---|---|---|---|
| Gene Name | FC | BH p-value | FC | BH p-value |
| CDC20 | 1.55 | 0.01 | 1.76 | 0.00 |
| CCNB2 | 1.55 | 0.01 | 1.66 | 0.00 |
| AURKA | 1.56 | 0.02 | 1.62 | 0.00 |
| BUB1 | 1.61 | 0.01 | 1.61 | 0.00 |
| NEK2 | 1.50 | 0.01 | 1.43 | 0.01 |
Biological Function Associated with Genes Found to be Dose-responsive to Alpha Particle Radiation in Keratinocytes
| Gene Name | Function |
|---|---|
| CDC20 | Anaphase Promoting Complex (APC) activator |
| CCNB2 | Cell cycle / regulation |
| AURKA | Protein kinase / mitotic process and regulation |
| BUB1 | Mitotic spindle checkpoint |
| NEK2 | Protein kinase / cell cycle |
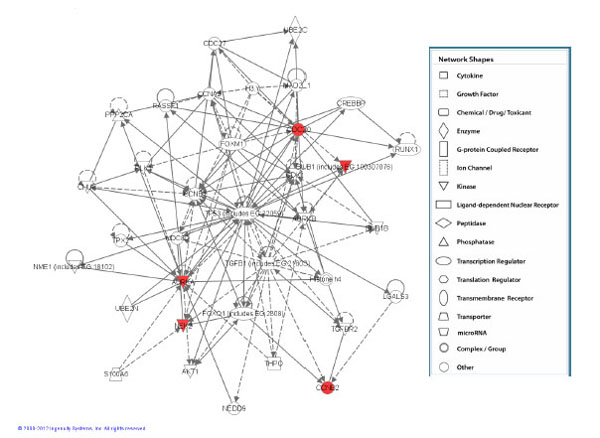
Network assembled from the 5 genes which were found to be significant at the medium and high doses. This network is shown to be clustered around TP-53 and TGFB signalling. Red represents upregulated genes.
A total of 68 genes were identified that were differentially expressed at the high dose (1.5 Gy) (Table 4a,b) of α-particle radiation when compared to control. Of these genes, 75% were upregulated and 25% were downregulated. Ninety three percent (93%) of the genes were exclusive to this dose of radiation and 7% of the total responding genes were also observed to be expressed at the medium (1.0 Gy) dose of radiation. The top expressing upregulated genes with fold changes greater than 1.8 fold were KIF20A, NEFM, CSN3, C11orf75, SLC16A9. Strongly repressed transcripts with FC≤-2 included C7orf10, HIST1H2BD, BMP6 and HIST1H2AC. A detailed functional analysis of the high expressing genes using a pathway analysis tool indicated that the top canonical pathways associated with this gene set were mitotic roles of polo-like kinase, cyclins and cell cycle regulation, chromosomal replication, ATM signaling and p53 signaling (Table 5). Nine genes were associated with these canonical pathways including KIF23, CDC20, CCNB2, CCNB1, CCNA2, MCM6, MCM2, TP53INP1 and BIRC5. Two top scoring pathways were obtained for this gene set. The first network was centered on TP-53 (Fig. 3A) signaling and the second top scoring network was clustered around ERK1 signaling (Fig. 3B).
Up-regulated Genes which were Statistically Significant (BH Corrected p Value < 0.05, FC ≥ 1.5) at the 1.5 Gy Dose of α-Radiation Sorted by Fold Change at this Dose. Other Dose Levels are Shown for Comparison
| 0.5 Gy | 1 Gy | 1.5 Gy | ||||
|---|---|---|---|---|---|---|
| Gene Name | FC | BH P-value | FC | BH P-value | FC | BH P-value |
| KIF20A | 1.16 | 1.00 | 1.57 | 0.24 | 2.08 | 0.00 |
| NEFM | 1.11 | 1.00 | 1.27 | 0.92 | 1.91 | 0.05 |
| CSN3 | -1.02 | 1.00 | 1.25 | 0.92 | 1.86 | 0.05 |
| C11orf75 | 1.05 | 1.00 | 1.33 | 0.81 | 1.82 | 0.01 |
| SLC16A9 | 1.10 | 1.00 | 1.37 | 0.54 | 1.81 | 0.00 |
| FAM83D | 1.17 | 1.00 | 1.43 | 0.64 | 1.77 | 0.01 |
| CDC20 | 1.15 | 1.00 | 1.55 | 0.01 | 1.76 | 0.00 |
| FUT4 | 1.34 | 1.00 | 1.50 | 0.43 | 1.74 | 0.01 |
| LMNB1 | -1.07 | 1.00 | 1.17 | 0.94 | 1.74 | 0.02 |
| SNORD96A | 1.12 | 1.00 | 1.26 | 0.89 | 1.73 | 0.03 |
| LOC646452 | 1.24 | 1.00 | 1.05 | 0.98 | 1.73 | 0.03 |
| CLDN11 | 1.12 | 1.00 | 1.20 | 0.92 | 1.72 | 0.03 |
| AURKB | 1.16 | 1.00 | 1.45 | 0.23 | 1.69 | 0.00 |
| MKI67 | 1.05 | 1.00 | 1.14 | 0.95 | 1.68 | 0.03 |
| CTNNAL1 | 1.14 | 1.00 | 1.30 | 0.78 | 1.68 | 0.01 |
| DLGAP5 | 1.09 | 1.00 | 1.52 | 0.32 | 1.68 | 0.02 |
| CCNA2 | 1.13 | 1.00 | 1.45 | 0.54 | 1.67 | 0.02 |
| HIST1H4C | 1.02 | 1.00 | 1.31 | 0.78 | 1.67 | 0.01 |
| SNORA32 | 1.61 | 0.53 | 1.27 | 0.84 | 1.67 | 0.02 |
| CCNB2 | 1.14 | 1.00 | 1.55 | 0.01 | 1.66 | 0.00 |
| CENPA | 1.05 | 1.00 | 1.49 | 0.28 | 1.64 | 0.01 |
| FAM64A | 1.13 | 1.00 | 1.37 | 0.30 | 1.63 | 0.00 |
| MCM6 | 1.02 | 1.00 | 1.15 | 0.94 | 1.63 | 0.03 |
| PSRC1 | 1.13 | 1.00 | 1.38 | 0.30 | 1.62 | 0.00 |
| CCDC58 | 1.05 | 1.00 | 1.36 | 0.61 | 1.62 | 0.01 |
| AURKA | 1.17 | 1.00 | 1.56 | 0.02 | 1.62 | 0.00 |
| MCM2 | 1.08 | 1.00 | 1.24 | 0.88 | 1.62 | 0.03 |
| TAF5 | 1.20 | 1.00 | 1.35 | 0.70 | 1.61 | 0.03 |
| FBXO5 | 1.11 | 1.00 | 1.24 | 0.88 | 1.61 | 0.03 |
| BUB1 | 1.21 | 1.00 | 1.61 | 0.01 | 1.61 | 0.00 |
| BIRC5 | 1.11 | 1.00 | 1.44 | 0.28 | 1.61 | 0.01 |
| NCAPG | 1.25 | 1.00 | 1.40 | 0.58 | 1.60 | 0.02 |
| NRG1 | 1.11 | 1.00 | 1.13 | 0.92 | 1.59 | 0.01 |
| CEP55 | 1.08 | 1.00 | 1.42 | 0.13 | 1.59 | 0.00 |
| KIF23 | 1.13 | 1.00 | 1.35 | 0.31 | 1.59 | 0.00 |
| ANLN | 1.17 | 1.00 | 1.56 | 0.09 | 1.58 | 0.01 |
| SPARC | 1.08 | 1.00 | 1.19 | 0.91 | 1.58 | 0.03 |
| CDCA3 | 1.18 | 1.00 | 1.36 | 0.69 | 1.57 | 0.04 |
| C16orf53 | 1.17 | 1.00 | 1.22 | 0.81 | 1.57 | 0.01 |
| CDKN3 | 1.08 | 1.00 | 1.46 | 0.16 | 1.56 | 0.01 |
| HJURP | 1.08 | 1.00 | 1.44 | 0.30 | 1.56 | 0.02 |
| CCNF | 1.07 | 1.00 | 1.38 | 0.55 | 1.54 | 0.02 |
| DLL1 | 1.01 | 1.00 | 1.13 | 0.93 | 1.53 | 0.02 |
| SAP30 | 1.11 | 1.00 | 1.23 | 0.86 | 1.53 | 0.03 |
| C13orf34 | 1.12 | 1.00 | 1.41 | 0.29 | 1.53 | 0.01 |
| SFRS10 | 1.18 | 1.00 | 1.31 | 0.56 | 1.53 | 0.01 |
| ASPM | 1.17 | 1.00 | 1.33 | 0.53 | 1.53 | 0.01 |
| CCNB1 | 1.27 | 1.00 | 1.56 | 0.13 | 1.53 | 0.03 |
| HMGB2 | 1.01 | 1.00 | 1.22 | 0.73 | 1.52 | 0.00 |
| IL7R | 1.09 | 1.00 | 1.20 | 0.88 | 1.50 | 0.04 |
Down-regulated Genes which were Statistically Significant (BH Corrected p Value < 0.05, FC ≥ 1.5) at the 1.5 Gy Dose of α-Radiation Sorted by Fold Change. Other Doses are Shown for Comparison
| 0.5 Gy | 1 Gy | 1.5 Gy | ||||
|---|---|---|---|---|---|---|
| Gene Name | FC | BH P-value | FC | BH P-value | FC | BH P-value |
| C7orf10 | 1.10 | 1.00 | -1.27 | 0.92 | -2.37 | 0.00 |
| HIST1H2BD | -1.06 | 1.00 | -1.42 | 0.61 | -2.03 | 0.00 |
| BMP6 | -1.10 | 1.00 | -1.37 | 0.84 | -2.01 | 0.01 |
| HIST1H2AC | -1.06 | 1.00 | -1.29 | 0.90 | -2.01 | 0.01 |
| INPP5D | -1.10 | 1.00 | -1.34 | 0.86 | -1.98 | 0.01 |
| TRIM22 | -1.09 | 1.00 | -1.49 | 0.61 | -1.83 | 0.02 |
| LOC100128208 | -1.14 | 1.00 | -1.01 | 0.99 | -1.83 | 0.03 |
| GSDMA | -1.20 | 1.00 | -1.36 | 0.62 | -1.72 | 0.01 |
| DAPK1 | -1.08 | 1.00 | -1.28 | 0.61 | -1.70 | 0.00 |
| MEG3 | 1.11 | 1.00 | -1.25 | 0.89 | -1.67 | 0.03 |
| SPATA18 | 1.01 | 1.00 | -1.20 | 0.89 | -1.66 | 0.01 |
| FERMT1 | 1.02 | 1.00 | -1.21 | 0.85 | -1.62 | 0.01 |
| TP53INP1 | 1.01 | 1.00 | -1.11 | 0.94 | -1.62 | 0.01 |
| HES2 | -1.04 | 1.00 | -1.20 | 0.89 | -1.62 | 0.02 |
| DLGAP2 | -1.59 | 0.53 | -1.33 | 0.70 | -1.61 | 0.02 |
| APOBEC3C | -1.11 | 1.00 | -1.34 | 0.70 | -1.56 | 0.04 |
| HIST1H2BK | -1.06 | 1.00 | -1.15 | 0.92 | -1.50 | 0.02 |
Terms Associated with the High Dose-responsive Gene List as Determined Through the Ingenuity Pathway Analysis (IPA) Bioinformatic Tools. The “Ratio” Heading Represents How Many Genes were Differentially Expressed in a Canonical Pathway Divided by the Number of Genes in the Pathway. The “Molecules” Heading Shows which Genes are Differentially Expressed within the Pathway
| Ingenuity Canonical Pathways | -log(p-value) | Ratio | Molecules |
|---|---|---|---|
| Mitotic Roles of Polo-Like Kinase | 4.26 | 6.15E-02 | KIF23,CDC20,CCNB2,CCNB1 |
| Cyclins and Cell Cycle Regulation | 2.62 | 3.37E-02 | CCNA2,CCNB2,CCNB1 |
| Cell Cycle Control of Chromosomal Replication | 2.43 | 6.67E-02 | MCM6,MCM2 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 2.02 | 4.08E-02 | CCNB2,CCNB1 |
| ATM Signaling | 1.88 | 3.7E-02 | CCNB2,CCNB1 |
| p53 Signaling | 1.41 | 2.11E-02 | TP53INP1,BIRC5 |
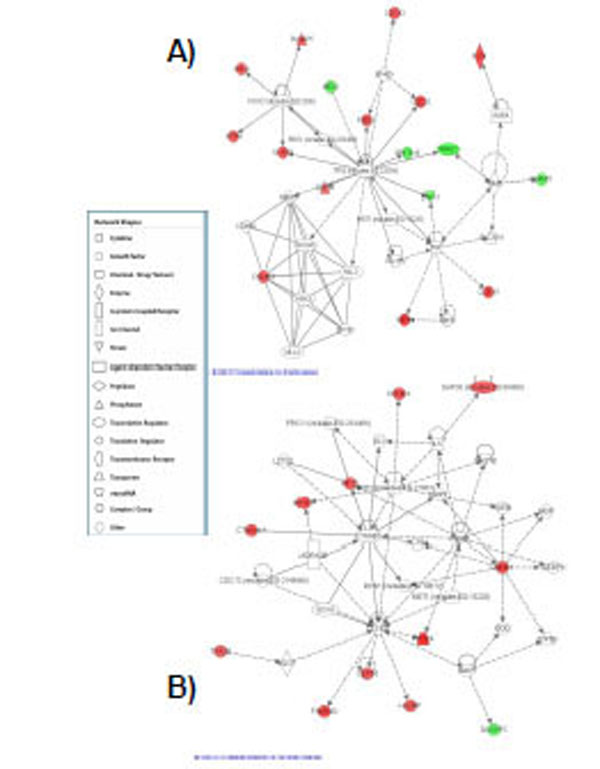
A) Highest scoring network assembled from the genes responsive at the highest dose of radiation tested. This network was centered on ERK1 signalling. Red represents upregulated genes and green represents downregulated genes. B) Network assembled from the genes responsive at the high dose and clustered around TP-53 signalling. Red represents upregulated genes and green represents downregulated genes.
Validation of Altered Expression of Radiation-Inducible Genes
For a selected few genes, for which validated primers were available, the observed microarrays results were verified using qRT-PCR. As shown in Table 6, all genes that exhibited a significant response at the high dose (1.5 Gy) of α-particle radiation 24 h post-exposure using microarray analysis were also observed to exhibit a similar trend using qRT-PCR. A comparable pattern of expression was also observed at the low dose (0.5 Gy) using the two methodologies. At the medium dose, although the fold changes were similar, the significance level did not always correspond. The five genes CDC20, CCNB2, AURKA, BUB1 and NEK2 that were shown to be significantly modulated at both the medium and high dose of radiation using microarray technology did express similar fold changes using qRT-PCR. However, these genes were not shown to be statistically significant with the exception of CDC20. Overall, these results suggest a comparable response between the two technologies, particularly at the high dose level.
qRT-PCR Validation of High Expressing Significant Genes Using Microarray Analysis. MA=microarray, FC=fold Change, BH=Benjamini-Hochberg Corrected p-value
| 0.5 Gy | 1.0 Gy | 1.5 Gy | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | MA FC | BH P-value | qPCR FC | P-value | MA FC | BH P-value | qPCR FC | P-value | MA FC | BH P-value | qPCR FC | P-value |
| KIF20A | 1.16 | 1.00 | 1.38 | 0.31 | 1.57 | 0.24 | 1.77 | 0.05 | 2.08 | 0.00 | 2.63 | 0.00 |
| NEFM | 1.11 | 1.00 | -1.24 | 0.30 | 1.27 | 0.92 | 1.25 | 0.47 | 1.91 | 0.05 | 2.63 | 0.00 |
| SLC16A9 | 1.10 | 1.00 | 1.27 | 0.23 | 1.37 | 0.54 | 1.44 | 0.17 | 1.81 | 0.00 | 2.75 | 0.01 |
| CDC20 | 1.15 | 1.00 | 1.24 | 0.26 | 1.55 | 0.01 | 1.77 | 0.00 | 1.76 | 0.00 | 2.06 | 0.01 |
| LMNB1 | -1.07 | 1.00 | 1.34 | 0.47 | 1.17 | 0.94 | 1.61 | 0.23 | 1.74 | 0.02 | 2.90 | 0.00 |
| CCNB2 | 1.14 | 1.00 | -1.13 | 0.57 | 1.55 | 0.01 | 1.24 | 0.16 | 1.66 | 0.00 | 1.52 | 0.02 |
| CENPA | 1.05 | 1.00 | -1.09 | 0.62 | 1.49 | 0.28 | 1.20 | 0.28 | 1.64 | 0.01 | 1.78 | 0.03 |
| AURKA | 1.17 | 1.00 | 1.11 | 0.58 | 1.56 | 0.02 | 1.73 | 0.22 | 1.62 | 0.00 | 1.74 | 0.02 |
| BUB1 | 1.21 | 1.00 | -1.01 | 0.84 | 1.61 | 0.01 | 1.10 | 0.63 | 1.61 | 0.00 | 1.47 | 0.02 |
| HMGB2 | 1.01 | 1.00 | 1.13 | 0.46 | 1.22 | 0.73 | 1.40 | 0.10 | 1.52 | 0.00 | 1.79 | 0.05 |
| C7orf10 | 1.10 | 1.00 | 1.24 | 0.28 | -1.27 | 0.92 | -1.16 | 0.41 | -2.37 | 0.00 | -1.26 | 0.21 |
| BMP6 | -1.10 | 1.00 | -1.16 | 0.70 | -1.37 | 0.84 | -1.62 | 0.09 | -2.01 | 0.01 | -5.13 | 0.00 |
| INPP5D | -1.10 | 1.00 | -1.16 | 0.68 | -1.34 | 0.86 | -1.56 | 0.15 | -1.98 | 0.01 | -3.04 | 0.00 |
| DAPK1 | -1.08 | 1.00 | 1.14 | 0.47 | -1.28 | 0.61 | -1.64 | 0.00 | -1.70 | 0.00 | -3.92 | 0.00 |
| MEG3 | 1.11 | 1.00 | 1.28 | 0.30 | -1.25 | 0.89 | -1.50 | 0.07 | -1.67 | 0.03 | -2.55 | 0.00 |
| SPATA18 | 1.01 | 1.00 | 1.08 | 0.58 | -1.20 | 0.89 | -1.16 | 0.49 | -1.66 | 0.01 | -1.80 | 0.01 |
| FERMT1 | 1.02 | 1.00 | 1.15 | 0.54 | -1.21 | 0.85 | -1.23 | 0.19 | -1.62 | 0.01 | -2.20 | 0.00 |
| TP53INP1 | 1.01 | 1.00 | 1.13 | 0.50 | -1.11 | 0.94 | -1.32 | 0.16 | -1.62 | 0.01 | -2.23 | 0.00 |
| NEK2 | 1.17 | 1.00 | -1.06 | 0.70 | 1.50 | 0.01 | 1.21 | 0.16 | 1.43 | 0.01 | 1.46 | 0.00 |
| GDF15 | -1.19 | 1.00 | -1.17 | 0.76 | -1.49 | 0.93 | -1.77 | 0.14 | -2.19 | 0.27 | -3.12 | 0.01 |
Bio-Plex Analysis
Supernatants from cells exposed to α-particle radiation and X-rays were assessed for the relative expression of 27 secreted cytokines over a time-course of 96 h. Analysis using two-way ANOVA (TIME*DOSE) revealed dose main effects (p<0.05) at each of the doses. Cytokines that were shown to be statistically significant at all three doses of α-particle radiation were plotted (PDGF-bb, IL-13 and RANTES) over the time-course of the experiment (Fig. 4). A similar analysis conducted for X-irradiated cells also revealed dose main effects (p<0.05). The expression of 6 cytokines (IP-10, IL-12, IL-10, RANTES, IL-5 and MIP-1a) was observed to be modulated for all three doses (Fig. 5). For both radiation types, the DOSE *TIME interaction did not reveal statistical significance at all interactions, however a clear biological trend was observed across the doses and time-points (Table 7).
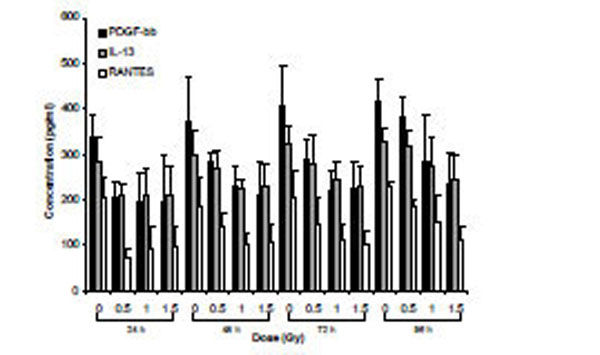
Secretomic analysis of cell supernatants exposed to α-particle radiation using Bio-Plex technology. Data are presented for cytokines that exhibited dose main effects using ANOVA.
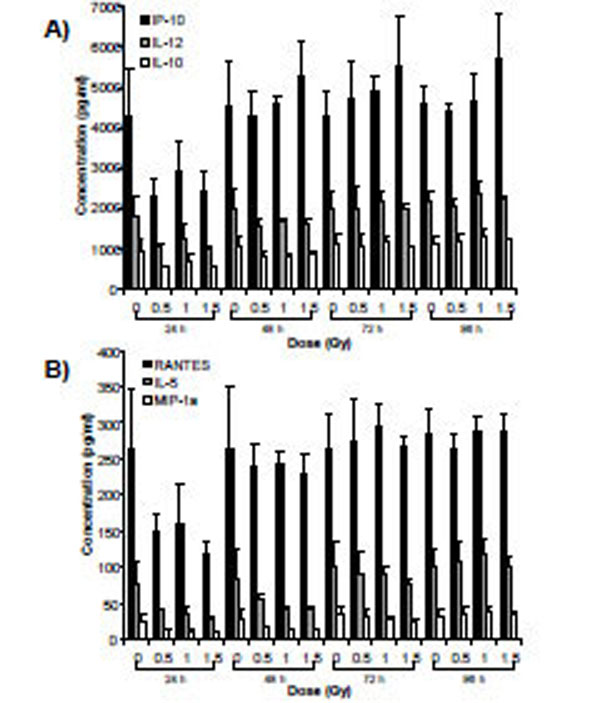
Secretomic analysis of cell supernatants exposed to X- radiation using Bio-Plex technology. Data are presented for cytokines that exhibited dose main effects using ANOVA. (a) high expression levels (b) moderate expression levels.
Statistical Significance of the Time, Dose and Time * dose Interactions for the Cytokines which were Found to Respond in a Dose Dependant Manner. Based on an n=5 Biological Replicates
| Time Interaction (p-value) | Dose Interaction (p-value) | Time x Dose Interaction (p-value) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 h | 72 h | 96 h | 0.5 Gy | 1.0 Gy | 1.5 Gy | 48 h x 0.5 Gy | 72 h x 0.5 Gy | 96 h x 0.5 Gy | 48 h x 1.0 Gy | 72 h x 1.0 Gy | 96 h x 1.0 Gy | 48 h x 1.5 Gy | 72 h x 1.5 Gy | 96 h x 1.5 Gy | ||
| Alpha | ||||||||||||||||
| PDGF-bb | 0.23 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.27 | 0.70 | 0.02 | 0.91 | 0.31 | 0.65 | 0.62 | 0.34 | 0.38 | |
| IL-13 | 0.48 | 0.04 | 0.02 | 0.03 | 0.03 | 0.03 | 0.13 | 0.33 | 0.04 | 0.95 | 0.81 | 0.59 | 0.84 | 0.44 | 0.65 | |
| RANTES | 0.28 | 0.98 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.23 | 0.38 | 0.20 | 0.23 | 0.76 | 0.68 | |
| X-ray | ||||||||||||||||
| IL-5 | 0.45 | 0.01 | 0.01 | 0.03 | 0.01 | 0.00 | 0.50 | 0.09 | 0.00 | 0.97 | 0.03 | 0.00 | 0.54 | 0.07 | 0.00 | |
| IL-10 | 0.29 | 0.04 | 0.03 | 0.00 | 0.01 | 0.00 | 0.09 | 0.00 | 0.00 | 0.40 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | |
| IL-12 | 0.22 | 0.14 | 0.01 | 0.00 | 0.01 | 0.00 | 0.08 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | |
| IP-10 | 0.45 | 0.89 | 0.37 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | |
| MIP-1a | 0.62 | 0.00 | 0.01 | 0.04 | 0.01 | 0.01 | 0.53 | 0.09 | 0.00 | 0.84 | 0.02 | 0.00 | 0.59 | 0.12 | 0.00 | |
| RANTES | 0.91 | 0.91 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
DISCUSSION
As the malicious use of radiation remains a concern, action plans which include forensic tools to potentially prevent and identify the perpetrators would be of benefit. Therefore, research and development into these areas are needed for new and improved methods to manage and mitigate nuclear/radiological threats. In this study, microarray technology alongside secretomics was employed to assess the biological effects of α-particles on dermal cells with the goal of identifying biomarkers of exposure. Gene and protein based markers could be used to create a bio-chips for identifying perpetrators that have handled special nuclear materials emitting α-particles
The skin is the first line of defense against foreign insults, and keratinocytes serve as a barrier between the living being and its environment. These cells play a role as immunomodulaters, and are responsible for secreting inhibitory cytokines in the absence of injury, and stimulating inflammation and activating antigen-presenting cells when there is a skin infection or injury (10). This chain of events can trigger subtle changes in the micro-environment of the cells leading to transcriptional and translational modifications in gene/protein expression, all of which can be monitored and potentially act as signature biomarkers of exposure [26]. In this study, keratinocytes were targeted for an initial screening to determine if α-particle radiation could elicit changes in the gene/protein micro-environment of the cell. Specifically, an evaluation of the response was assessed for intensity, dose range of detection and the specificity through a comparative assessment using an alternate radiation type i.e X-rays. To be strong biomarkers, the gene response should be pronounced (>2 fold) for detection using bio-chip technology, specific and sustained over a range of doses.
Screening of 47 000 gene probes for differential expression at three doses of α-particle radiation led to limited transcript modulations, particularly at the lower doses that were tested. The lowest dose of radiation (0.5 Gy) did not elicit changes in gene expression using stringent statistical methods. The less stringent statistical methods that did not employ the BH false discovery correction yielded some interesting gene responses. However, given that the gene responses were not expressed at the higher doses and had relatively low fold change, they may not represent ideal biomarkers. The moderate dose of radiation (1.0 Gy) caused differential expression in the levels of 5 genes, all associated with mitotic processes. These 5 genes were also shown to be expressed at the higher dose of radiation (1.5 Gy), however of the 5 genes, only CDC20 validated using alternate technology (qRT-PCR) at both doses. CDC20 was shown to be upregulated by 1.5 fold at the medium dose and 1.7 fold at the higher dose of radiation relative to the control treatment group. With such low levels of modulation, it would not lend itself to be a strong candidate biomarker. However, it is of interest that all 5 genes were associated with mitotic processes centered on TP-53 and TGFB signaling, as this implicates an important role for the mitotic checkpoint and mitosis canonical pathway for focused pathway array studies.
The highest dose of of α-particle radiation (1.5 Gy) did elicit a strong gene response. Keratinocytes exposed to 1.5 Gy of α-particle radiation caused the differential expression of 67 genes relative to the control treatment group, among these genes a selected few had fold expression of 2 or higher. The canonical pathways associated with these genes were clustered around cell cycle control /chromosomal replication/p53 signaling and ATM signaling. Additionally, a number of these targets validated using alternate technology and were not expressed in X-irradiated cells. Among the upregulated genes, KIF20A and NEFM may be of interest for further validation studies and among the downregulated genes C7orf10, HIST1H2BD, BMP6, and HIST1H2AC may warrant further studies to assess if these transcripts are modulated in a temporal sequence and at higher doses of α-particle radiation.
Secretomic analysis in the expression of 27 cytokines revealed a limited response to radiation insults. X-irradiation cells modulated the expression of 6 cytokines, IP-10, IL-12, IL-10, RANTES, IL-5 and MIP-1a. Alpha-particle irradiated cells had modulated the expression of three cytokines (PDGF-bb, IL-13 and RANTES) over the doses examined. RANTES was commonly expressed between the two radiation types. PDGF-bb and IL-13 were exclusive to α-irradiated cells and expression levels of these cytokines were shown to be downregulated relative to the control treatment groups. Furthermore, this expression was sustained over a 4 day time-course. Although these cytokines may be of interest, given their decrease expression levels they may not lend themselves to being strong biomarkers of radiation exposure. The overall limited response in pro- and anti-inflammatory cytokine secretion implies that keratinocytes may not be strong expressors of these types of cytokines. For future studies, other sub-sets of secretory proteins may be assessed for differential expression.
CONCLUSION
In conclusion, an initial screening using secretomic/ genomic technology of skin cells exposed to α-particles resulted in a limited response. The responding genes were clustered around pathways related to cell cycle control with TP-53 and TGF signalling. One gene, CDC20, was shown to be unique to α-particle irradiated cells and was validated to be dose-responsive. Several genes were differentially expressed at only the high dose of radiation; of particular interest are those transcripts with 2 fold regulation (KIF20A, NEFM, C7orf10, HIST1H2BD, BMP6, and HIST1H2AC). While gene-based profiling of the skin yielded differential regulation results, further validations of these responses should be conducted to determine their specificity and robustness. Secretomic analysis using Bio-plex technology also resulted in a limited number of responding cytokines. Although skin is a likely place of contamination, blood may also serve as a useful biological end-point for assay. Some α-particle emitting isotopes have been shown to clear from the lung to the blood in a short amount of time, and isotopes can also enter circulation through a compromised dermal layer or wounds. Future studies will explore other endpoints including focused pathway array profiling and metabolomics using blood drawn from healthy individuals
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENTS
This work was supported by the Canadian Centre for Security Studies and funding was provided by the Chemical, Biological, Radiological-Nuclear, and Explosives (CBRNE) Research and Technology Initiative (CRTI). The authors wish to thank Ms. Kate Daniel for her technical expertise and assistance with the cytokine assays and analysis.
ABBREVIATIONS
REFERENCES
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]


