All published articles of this journal are available on ScienceDirect.
Study of the Interaction of Zinc Cation with Azithromycin and its Significance in the COVID-19 Treatment: A Molecular Approach
Abstract
Introduction:
On account of the current COVID-19 pandemic, we have explored the importance of azithromycin and zinc in the treatment of the coronavirus disease by studying the interaction between the cation Zn++ and azithromycin with the tools of the semi-empirical quantum mechanics PM3 method.
Methods:
By this approach, the niche in which Zn++ is located was determined. Zn++ creates a strong clastic binding between an amine and a hydroxyl group located on the amino-hexose side-chain. Such an interaction serves as a shuttle and allows zinc cation to invade endocellular structures.
Results:
In this triple collaborative association, the role of hydroxychloroquine would be more that of a chaotropic agent at plasmic membranes, which facilitates access to the azithromycin-Zn++ equipage into key internal compartments.
Conclusion:
Finally, we show that both azithromycin and Zn++ are susceptible to play a direct role against the replication and the assembly of SARS-CoV-2 particles.
1. INTRODUCTION
With the advent of the COVID-19 pandemic, one of the proposed current treatments consists of a tritherapy associating azithromycin, hydroxychloroquine and zinc (II) sulfate. In an effort to study more in-depth the rationale, we explored the interaction of the cation Zn++ with azithromycin. Azithromycin (Fig. 1) occupies a leading position as a major macrolide antibiotic within the group of azalides. It is used to treat respiratory tract, soft tissue, and genitourinary infections [1-3]. Azithromycin structurally differs from erythromycin (Fig. 1) by the insertion of a nitrogen atom into the lactone ring of erythromycin A, making this lactone ring a 15-atom ring. This modification renders this macrolide more stable in an acid medium and increases both plasma half-life and oral bioavailability. Indeed, while azithromycin does not conform to Lipinsky’s and Weber’s rules of druggability (MW 749 Da, 15 heteroatoms) [4], oral bioavailability is in the order of 40% [5]. However, this favorable parameter is hampered by co-administration with multivalent metal cations such as magnesium and aluminium [6]. Previous works performed by Sher et al. [7] on the interaction of azithromycin with copper (II) ion (III) and Rjoob et al. [8] also support the affinity of azithromycin for metal cations. All these electropositive species belong to the group of Lewis acids and interact strongly with Lewis bases, which are characterized by the existence of available lone pairs decorating these molecules. The presence of 14 such heteroatoms within its structure allows to rationalize the above behavior. Such Lewis salts have been synthesized recently by Arayne et al. [9] and characterized by spectroscopy and elemental analysis. This study shows that these salts are structured in fact around a bidentate complex geometry with the metal cation occupying a central position.
Within the context of the dramatic emergence of COVID-19 by the end of 2019, Prof. Raoult from Marseille, France and his research group has proposed a tritherapy associating azithromycin, hydroxychloroquine and zinc (II) sulfate [10]. In view of the Pakistan’s publication [9], in an effort to shed light on the charisma of zinc (II) cations in this therapeutic association, a molecular modeling study involving quantum mechanics tools has been implemented. The purpose of this paper is to describe our approach and results along this line.
2. EXPERIMENTAL PROCEDURE
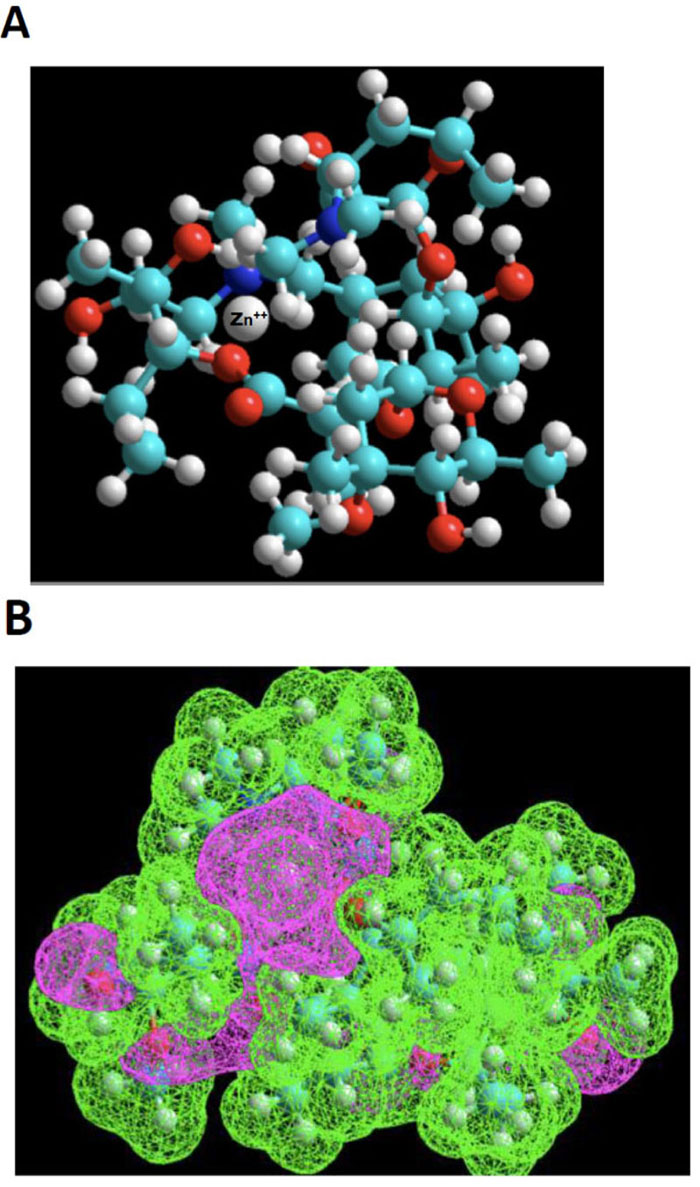
Azithromycin 3-D structure was built starting from the molfile obtained from the data bank of the European Bioinformatics Institute (EMBL-EBI). MDL Molfile is a file format containing information about the atoms, bonds, connectivity and coordinates of a molecule, and this format is readable by the HyperChem package. The resulting 3D-structure was extensively energy-minimized first using molecular mechanics OPLS (Optimized Potentials for Liquid Simulations) force field and then subjected to a molecular dynamics trajectory target temperature 310 K) in order to sample conformers every 10 psec for a total of 250 psec. Using semi-empirical quantum mechanics PM3, resulting conformers were then energy-minimized down to a gradient of 0.01 (~5000 iterations). Indeed, PM3 (for Parametric Method 3) derived from AM1 is a semi-empirical method for the quantum calculation of molecular electronic structure in computational chemistry. It is based on the Neglect of Differential Diatomic Overlap integral approximation. For this purpose, we employed the Polak-Ribiere Nonlinear conjugate gradient method. The resulting low-energy conformer structure was then empirically exposed to a zinc (II) cation probing different loci to search for potential interactions with Lewis base sites. Most attempts result in quick repulsion of the zinc atom, which is expelled after just a few iterations at a long distance from azithromycin. When instead of using the energy minimization tool to search for suitable niches for Zn (II) cation, and instead, the “transition state” tool was used, we were finally met with some success. The zinc cation would stay in the vicinity of azithromycin’s cortex and explore a large area before finding an adequate docking site. This approach showed that this “voyage” implied considerable deformation of azithromycin’s skeleton. One such exploration led to a rather successful “landing”, as depicted in the ball and stick model shown in Fig. (2A).
Subsequent after PM3 calculations, due to excessively huge time-consuming computation times, a limited number of calculations (essentially energy assessments and energy minimizations) were also carried out using “Ab Initio” Quantum Mechanics at the 3-21G level.
3. RESULTS AND DISCUSSION
3.1. Molecular Modeling
The zinc element, strangely enough for its atomic mass (65.4 mass units), possesses a relatively small Van der Waals (VdW) radius (139 pm) compared to protium (120 pm) or carbon (170 pm). Considering the double positive charge of this metallic ion, Zn++ irradiates at long distance (~10 times its VdW radius) a considerable electromagnetic field and, therefore, can be recognized from far by Lewis base sites. The positive charge is indeed localized in a small volume. Zn++ is therefore, also classified among the strong Lewis acid and is often regarded as oxophilic in the same way as Li+ [11]. On the other hand, however, this electroattractive effect is negatively counteracted by the overwhelming shielding of the positively charged hydrogen cortex surrounding azithromycin (72 at the total for C38H72N2O12).
Based on the above premise, it is therefore not surprising that our “landing” exploration attempts were initially met with poor success. However, using the “transition state” tool, we were able to observe the dynamic behavior of Zn++ in its exploration of a large topological territory, which reflects its intrinsic affinity. Even in a volume of space very electron-rich, for instance, in the neighborhood of the two ose side-chains, this exploration was highly time-consuming. The reason for this can be found in the narrow access to the gate between the pincer mouth of the twin sugar-like moieties. The successful candidate exhibits a strong clastic binding with a hydroxyl group and the N-dimethylamino moiety, as shown in Fig. (2B), estimated by thermodynamic cycling in the order of 30 kcal/mol.
The results of our molecular simulations are somehow in harmony with the findings of the pioneering work of Arayne et al. [9] because the same two functional groups are in effect involved in Zn++ recognition, and our structure is also coherent with their NMR work. It is also worth noting that Zn++ is in the vicinity of two additional oxygen atoms. The bidentate structure proposed by the Pakistani group is based on evidence arising from convincing elemental analysis data [9]. However, these highly organized assemblies are obtained after a long process of crystallization (authors mentioned several weeks). The slow progress of crystal nucleation and growth is likely to be due to a negative entropy factor. Therefore, the first recognition process allowing recruitment of Zn++ by single azithromycin is absolutely mandatory further to recruit a second macrolide molecule in the crystal cell. In summary, the first recognition is relatively fast considering the high mutual affinity, the second one to assemble the bidentate system is rather slow, and the two steps are reversible. In other words, the 1:1 complex is the kinetic product, while the 1:2 complex is the thermodynamic product.
Our depiction of the 1:1 Zn++-azithromycin complex is likely to be the same under in vivo conditions. In the tritherapy treatment of COVID-19 as proposed and successfully tested by Prof. Raoult’s group in Marseille (France), all three active principles are administered together. As a consequence, initially, there exists a high local concentration of Zn++ in the presence of azithromycin, and therefore, the production of the 1:1 complex should take place rapidly. That there is, therefore, a synergy between both of them makes no doubt, but is there another asset? An attractive hypothesis would be that azithromycin plays as an ionophore the role of Trojan horse for Zn++ and would allow this highly polar hydrophilic Lewis acid to invade cellular compartments normally poorly accessible to Zn++. It is interesting to note that a zinc:erythromycin complex had been previously described [12, 13]. This observation suggests that both first-generation and second-generation macrolides (namely, erythromycin and azithromycin, respectively) are equally capable of forming a Zn++-antibiotic complex.
Beyond this, Zn++ ionophore role of azithromycin, one may wonder what is the part of hydroxychloroquine in this interplay? While all three individuals have in their own a crucial intrinsic role, another attractive hypothesis opens it up: hydroxychloroquine may play a role of the chaotropic agent at the level of the plasmic cell membrane and also at other endocellular barriers. Indeed, the quinoleine aromatic nucleus is a planar template fitted with a highly mobile side-chain (with important segmental and rotational freedom) at physiological temperature. As soon as the plasmic cell membrane loses its local coherence, the conditions are optimal for giving access to the Zn++-azithromycin equipage into the normally forbidden territories.
Even though a controversial issue [14] has recently emerged about the role of hydroxychloroquine in the aforementioned tritherapy, the fact that all three active principles are administered together at the onset of COVID-19, upon the first manifestation among the most common symptoms (fatigue, fever, dry cough, headache, sore throat, muscle pain or shortness of breath), is likely to explain why the treatment of COVID-19 by Prof. Raoult’s group in Marseille (France) was successful in these specific conditions [10]. It remains that both hydroxychloroquine and azithromycin might behave as zinc transporters. In this view, the administration of the Raoult’s therapy without chloroquine might have similar positive effects without adverse side-effects of chloroquine treatment.
3.2. Effect of Azithromycin and Zn++ on the Replication of SARS-CoV-2 and on the Assembly of Viral Particles
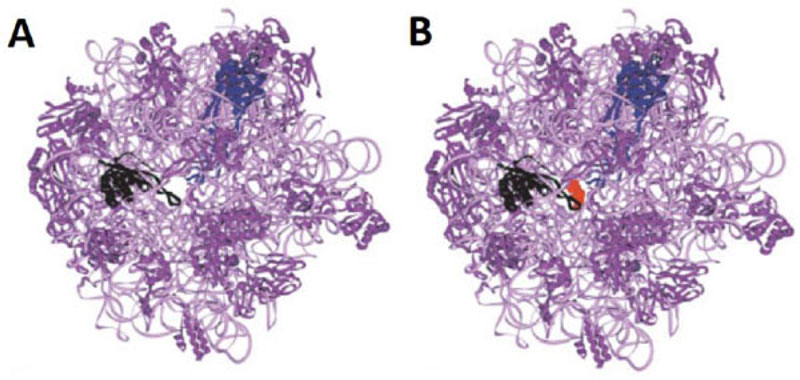
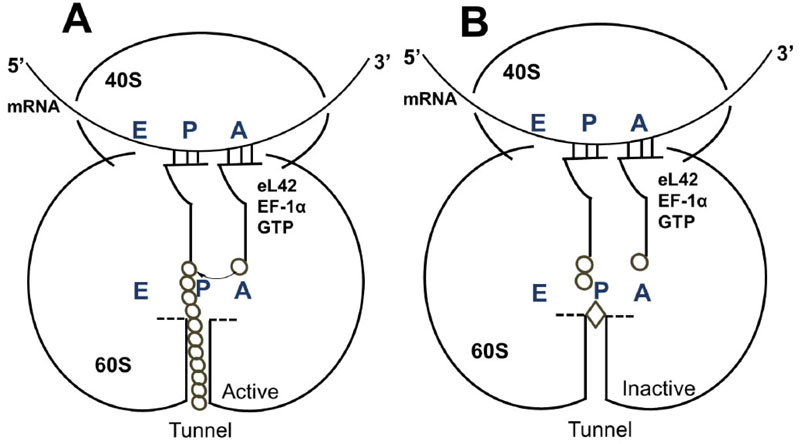
It is reasonable to consider that, in a host cell infected by the SARS-CoV-2 virus [15], the main target for the therapeutically useful drugs is the human ribosome, followed by individual proteins and enzymes of the virus [15], which are also synthesized by the ribosome itself. In fact, with the exception of the RNA genome that encodes all the viral proteins, the SARS-CoV-2 virus contains neither any major molecular material nor any protein synthesis machinery to produce the viral proteins and enzymes essential to its replication and/or to the assembly of viral particles. Among the viral individual proteins and enzymes targeted by antiviral drugs is the SARS-CoV-2 RNA replicase common to all RNA-containing viruses without any DNA stage [15]. It is an RNA-dependent RNA polymerase (RdRp), which catalyzes the synthesis of the RNA strand complementary to the viral RNA genome as a template [15]. Thus, RdRp is the core enzyme of the viral RNA-synthesizing machinery responsible for the replicative cycle of SARS-CoV-2, and as such, represents a key target for antiviral drug development [15]. In accordance with this observation, it had been previously demonstrated that Zn++ inhibits coronavirus RdRp in vitro, while zinc ionophores block the replication of the virus in cell culture [16]. Thus, on the one hand, it is tempting to propose that the azithromycin-Zn++ complex would represent the provider of Zn++ to the infected human host cells in order to inhibit the viral RdRp and prevent the replication of SARS-CoV-2. On the other hand, one should wonder if, instead of targeting RdRp with antiviral drugs, it is not preferable to use small-molecule inhibitors of the human ribosome in order to prevent global protein synthesis of all the viral proteins and enzymes. Interestingly, antibiotics are the most common small-molecule inhibitors of the ribosome, among which azithromycin is particularly suitable for the following reason: the binding site of the antibiotics of the macrolides class (including azithromycin) is located at the upper part of the ribosomal protein exit tunnel that is a universal feature of the ribosomes in all kingdoms of life [17-22]. Therefore, this class of antibiotics is expected to bind to all kinds of ribosomes, the 70S as well as the 80S type, with comparable affinity. Figs. (3A & B) shows the protein exit tunnel in the crystallographic structure of the 50S ribosomal subunit of Haloarcula marismortui [23], in the absence or the presence of a macrolide (namely carbomycin). Whether or not azithromycin binds alone or as a Zn++-azithromycin complex to the protein exit tunnel of the ribosome needs to be clarified. In addition, the schematic of the binding site of azithromycin on human 80S ribosomes is shown in Fig. (4). It is seen that azithromycin and the macrolides inhibit protein synthesis by interfering with the progression of the nascent peptide into the protein exit tunnel (Fig. 4A & B) [19-22]. In this respect, it is well documented that azithromycin is endowed with multiple anti-infectious properties, including antiviral ones. For instance, azithromycin is known as the gold standard in the treatment of papillomavirus in dogs [24]. It is interesting to note that Zn++ had been previously shown to inhibit translation initiation [25]. This observation would suggest that not only the zinc ion is an effective inhibitor of RdRp, but it is also capable of preventing the whole ribosome from synthesizing the RNA-synthesizing machinery responsible for the replicative cycle of SARS-CoV-2. Other roles of zinc include impairment of the replication of RNA viruses, by interfering with correct proteolytic processing of viral polyproteins, improvement of the antiviral immunity through up-regulation of interferon α production hence increased antiviral activity, anti-inflammatory activity, etc. [26]. Altogether, these observations would suggest that Zn++ may be considered as a strong antiviral agent in COVID-19 treatment. Another potential action of azithromycin (or erythromycin) on the ribosomes of the human host cells infected by SARS-CoV-2 is that, as discussed above, these antibiotics are capable of forming a complex with the zinc atom bound to the N-terminal region of the large subunit ribosomal protein eL42 (formerly L42A or L42AB in yeast, L36a or L36a-like in human, or L44e in archaea) ((Fig.5) and [27]). This newly discovered ribosomal protein was shown to contribute actively and directly to the catalytic activity of the ribosome, at the elongation step of translation, through interactions with the incoming A-site bound aminoacyl-tRNA. Given that, it is widely accepted that the zinc finger domains are essential to the nucleic acid: protein interactions and/or to maintain the 3-D structure of zinc-containing proteins, it is conceivable that azithromycin (or erythromycin) capture the zinc from the catalytic ribosomal protein eL42, leading to the inactivation of the 80S ribosomes in human host cells infected by SARS-CoV-2 upon treatment with these antibiotics [28, 29]. Altogether, these results would suggest that the Zn++-azithromycin complex can be considered as a multi-hit compound against the COVID-19.

CONCLUSION
In this paper, we provide evidence-based on quantum mechanics molecular simulations for the molecular events surrounding the interaction of Zn++, a key ingredient in the tritherapy proposed and clinically tested by Prof. Raoult’s team in Marseille (France). The data collected at the atomic level reinforce the role of azithromycin as an ionophore of Zn++. Taking into account the controversial issue that has recently emerged about the role of hydroxychloroquine, and considering that both hydroxychloroquine and azithromycin might behave as zinc transporters, it might be preferable to administer the Raoult’s therapy without hydroxychloroquine in order to avoid its adverse side-effects. Finally, the fact that both first-generation and second-generation macrolides (namely, erythromycin and azithromycin, respectively) are equally capable of forming a Zn++-antibiotic complex makes it possible to choose the antibiotic which has less side-effects for the treatment of the coronavirus disease.
LIST OF ABBREVIATIONS
The Abbreviations Used Are:
| rp | = ribosomal protein |
| eL42 | = eukaryal or archaeal large subunit ribosomal protein eL42 |
| RdRp | = RNA-dependent RNA polymerase. |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We are grateful to Prof. Philip S. Portoghese (University of Minnesota, Twin Cities, MN, USA) for long standing scientific exchanges and Dr. Giovambattista Scarfone (Cardiology Center Bianco, Italy) for relevant inspirational conversations. We gratefully thank Prof. Fernand Gbaguidi and Dr. Urbain C. Kassehin for their constant interest to this work, and for fruitful discussions.


