All published articles of this journal are available on ScienceDirect.
Purification and Biochemical Characterization of Cysteine Protease from Baby Kiwi (Actinidia arguta)
Abstract
Background:
It has recently been reported that the fruit, stems and leaves of Actinidia arguta have various potential health effects including an antioxidant effect, anticancer effect, anti-allergic effect and α-glucosidase inhibitory effect. However, little is known about the biochemical properties of cysteine protease in the fruit juice of A. arguta.
Methods:
Ion exchange chromatography to purify the cysteine protease from the fruit juice of A. arguta, and some synthetic substrates to determinate the enzyme activity were used.
Results:
Cysteine protease was purified to homogeneity from A. arguta fruit juice by ion exchange chromatography. The molecular weight of the purified enzyme was calculated to be approximately 25,500 by SDS-PAGE in the presence of β-ME. The enzyme rapidly hydrolyzed the substrate Z-Leu-Arg-MCA and moderately hydrolyzed other substrates including Boc-Val-Leu-Lys-MCA, Z-Val-Val-Arg-MCA and Z-Phe-Arg-MCA. Kinetic parameters for these four substrates were determined. The Km, Vmax, Kcat and Kcat/Km values for Z-Leu-Arg-MCA, the most preferentially cleaved by the enzyme, were 100 μM, 63.8 μmoles/mg/min, 27.26 sec-1 and 0.2726 sec-1μM-1, respectively. Furthermore, the activity of the enzyme was strongly inhibited by inhibitors including antipain, leupeptin, E-64, E-64c, kinin-free-LMW kininogen and cystatin C. Those biochemical data indicated that the enzyme was a cysteine protease. The amino acid sequence of the first 21 residues of cysteine protease purified from Actinidia arguta was Val1-Leu-Pro-Asp-Tyr5-Val-Asp-Trp-Arg-Ser10-Ala-Gly-Ala-Val-Val15-Asp-Ile-Lys-Ser-Qln20-Gly. This sequence showed high homology to the sequences of actinidin from Acinidia deliciosa (95.0%) and actinidin from Actinidia eriantha (90%). These three cysteine proteases were thought to be common allied species.
Conclusion:
The biochemical properties of the enzyme purified from A. arguta fruit juice were determined. These basic data are expected to contribute to the maintenance and improvement of human health as well as to the promotion of protein digestion and absorption through its proteolytic functions.
1. INTRODUCTION
Actinidia arguta Planch., known as baby kiwi, hardy kiwi or arctic kiwi in English and Sarunashi in Japanese, is a vine tree that is natively widespread in Northern Asia including Russian Siberia, Northern China, Korea, Japan [1] and also in southern Asia including Vietnam, Peninsular Malaysia and New Guinea [2].
A. arguta is an edible small fruit-producing perennial vine belonging to the Actinidia genus in the Actinidiaceae family and produces a small fruit that is rich in anthocyanins, catechins, chlorophyll, β-carotene, vitamin C, lutein, several minerals (potassium, calcium and zinc) and other polyphenols [3].
It has recently been reported that the fruit, stems and leaves of A. arguta have various potential health effects including an antioxidant effect [4-6], antiproliferative effect [4], anticancer property [7], antimutagenesis effect [8], anti-allergic effect [9] and α-glucosidase inhibitory effect [9, 10]. Thus, the properties of A. arguta are thought to be beneficial for human health.
Furthermore, the proteolytic properties of actinidin, bromelain, ficin and papain, which have been identified in kiwifruit [11, 12], pineapple [13], figs [14] and papaya [15], have been extensively studied as good meat tenderizers [12-16]. All of these proteases have been confirmed to be cysteine proteases [EC 3.4.22.-]. They are categorized into clan CA and family C1 (papain family) (see Merops: https://www.merops.ac.uk.
The cDNA structure and amino acid sequence derived from cDNA structures of cysteine protease from A. arguta have been reported [17], and the crystallographical structure of the enzyme has also been determined [18]. However, little is known about the biochemical and physiological properties of cysteine protease in the fruit juice of A. arguta [19].
In this paper, we describe a simple procedure for the isolation of cysteine protease from baby kiwi (A. arguta) fruit juice and we also report the biochemical and physiological properties including molecular weight, kinetic parameters and N-terminal amino acid sequence.
2. MATERIALS AND METHODS
2.1. Materials
Frozen fruit samples of baby kiwi (A. arguta) harvested in the vicinity of Sapporo City (Hokkaido, Japan) that had been ripened for 2 days at room temperature were stored at -30°C until use.
Silver vine (Actinidia polygama) was also harvested in the vicinity of Kushiro City (Hokkaido, Japan), and both green kiwi (Actinidia deliciosa cv. Hayward) and gold kiwi (Actinidia chinensis cv. Hort16A) fruits were purchased from a city market.
Protease inhibitors including E-64, and antipain and fluorogenic peptide substrates including Z-Leu-Arg-MCA and Z-Phe-Arg-MCA were purchased from Peptide Institute (Osaka, Japan). Bio-Safe Coomassie G-250, Quick startTM Bradford 1x dye solution, Mini-Protean TGX precast gels (any kD) and UNOspheare Q were obtained from Bio-Rad Laboratories (Hercules, CA, USA).
All other chemicals were of analytical grade.
2.2. Assay of Cysteine Protease Activity
Enzyme activity was measured by fluorometrical number (excitation, 360 nm; emission, 460 nm) of liberation of AMC at 37°C for 10 min in a mixture containing 10 μl of 10 mM of the substrate Z-Leu-Arg-MCA, 100 μl of 0.5 M Na-phosphate buffer, pH 7.0, 10~20 μl of diluted enzyme solution and Mili Q water (18 mΩ) in a total volume of 1 ml. Liberated AMC was determined with the VersaFluor Flurometer system (Bio-Rad, Hercules, CA, USA). One unit of activity was defined as the amount of enzyme that hydrolyzes 1 μmole of the substrate per minute.
2.3. Protein Quantitation
To determined protein concentrations, absorbance at 280 nm and/or a Protein assay kit (Bradford’s method [20]) (Bio-Rad Laboratories, Hercules, CA, USA) were used. The samples were analyzed in a Shimazu UV-1850 UV-VIS spectrophotometer (Kyoto, Japan).
2.4. Polyacrylamide Gel Electrophoresis and Molecular Weight Determination
Slab gel electrophoresis was carried out on polyacrylamide gels (Mini-protean TGX precast gels (any kD or 4~20%)) obtained from Bio-Rad Laboratories (Bio-Rad, Hercules, CA, USA) by the method of Laemmli [21]. Proteins in the gel were stained with Bio-Safe Coomassie Brilliant Blue G-250 (Bio-Rad Laboratories, Hercules, CA, USA).
2.5. Molecular Weight Determination
The molecular weight of the enzyme was determined by SDS-PAGE. As the standard protein, precision plus proteinTM prestained standard (Mr 10 ~ 250 kDa) obtained from Bio-Rad Laboratories (Hercules, CA, USA) was used.
2.6. Purification of Cysteine Protease from Actinidia Arguta
Step 1. Extraction of Protease from Baby Kiwi Fruits and Dialysis
Baby kiwi fruits (~250 g) were chopped into small pieces after peeling off the skin, and one volume of cold 40 mM Tris–HCl buffer, pH 8.0, containing 10 mM β-ME was added. The solution was centrifuged at 12,500 x g for 45 min, and then the supernatant obtained was dialyzed overnight against 20 mM Tris-HCl buffer, pH 8.0, containing 10 mM β-ME. To remove the precipitate, the supernatant was centrifuged again at 12,500 x g for 45 min.
Step 2. UNO sphere Q Column Chromatography
The dialysate was applied at a flow rate of 1.0 ml/min to a UNOsphere Q column (bed volume, 20 ml) pre-equilibrated with 20 mM Tris-HCl buffer, pH 8.0, containing 10 mM β-ME. The column was washed extensively with the same equilibration buffer, and then a linear gradient was formed with 100 ml of the same buffer and 100 ml of the same buffer containing 0.45 M NaCl. Fractions containing cysteine protease activity were collected. The solution was immediately used in subsequent experiments.
2.6.1. Kinetic Analysis
The concentration of an inhibitor that gave 50% inhibition (IC50) was determined through a series of assays with a fixed substrate concentration of 100 μM but with various inhibitor concentrations.
To determine Km and Vmax values, double-reciprocal plots (Lineweaver-Burk plots) were utilized.
2.6.2. N-Terminal Amino Acid Sequencing
Purified cysteine protease (~ 20 μg: ~ 0.77 pmol) was subjected to electrophoresis in 15% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories). The membrane was stained with 0.1% Coomassie Brilliant Blue R-250 in 10% methanol-7% acetic acid. The protein band to be examined was cut out, washed with 50% methanol, and then sequenced by automated Edman degradation using an Applied Biosystems Model 492 cLC (ABI, Foster City, CA, USA). A protein sequencer equipped with an online phenylthiohydantoin analyzer (ABI 120A analyzer, Foster City, CA, USA) was used.
3. RESULTS AND DISCUSSION
3.1. Activation of Cysteine Protease by β-ME
The enzyme from A. arguta fruit juice in the absence of β-ME showed very low activity using the substrate Z-Leu-Arg-MCA. However, in the presence of 5~15 mM β-ME, the enzyme showed about 160~220-fold higher activity than the activity in the absence of β-ME (Fig. 1). These results indicated that the enzyme exists as a precursor form in the fruit juice and that the enzyme is activated by sulfhydryl reagents such as β-ME and cysteine [22, 23]. Accordingly, 10 mM β-ME was added to the assay mixture and purification buffer (Tris-HCl buffer, pH 8.0) in subsequent experiments.
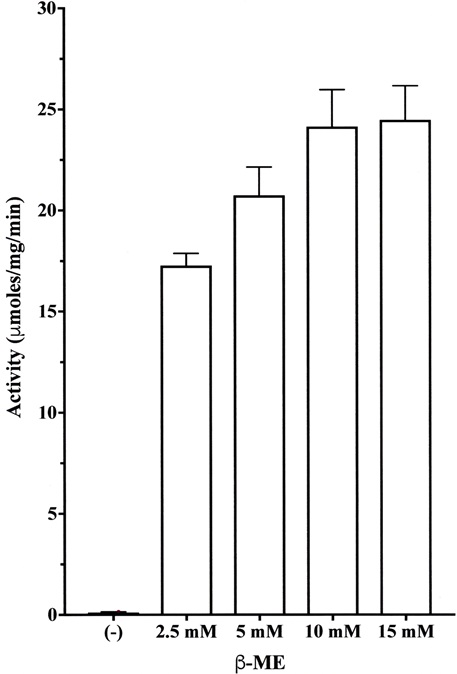
Each value is the mean ± SE from triplicate experiments.
3.2. Comparison of Specific Activities Among A. arguta, A. delisiosa, A. chinensis and A. polygama
The enzyme activities of fruit juices extracted from A. arguta, A. delisiosa, A. chinensis and A. polygama were determined in the presence of 10 mM β-ME using the substrate Z-Leu-Arg-MCA. The activity of A. arguta fruit juice showed the highest activity (Table 1). The order of their relative activities was A. arguta (16.97 μmoles/mg/min, 100%) > A. delisiosa (4.70 μmoles/mg/min, 45.4%) > A. polygama (0.83 μmoles/mg/min, 4.89%) > A. chinensis (0.83 μmoles/mg/min, 0.0015%).
3.3. Enzyme Purification
Cysteine protease from A. arguta was purified by chromatography on a UNOsphere Q column (Fig. 2). Table 2 shows a typical procedure for purification of the enzyme. Finally, the cysteine protease from A. arguta was purified approximately 2.12-fold with a 62.1% yield over dialyzed A. arguta homogenate. The enzyme in the final step gave a single band on SDS-PAGE in the presence of β-ME (Fig. 2). The overall yield of cysteine protease from 100 g of A. arguta was approximately 13.3 mg.
| Specific Activity (μmoles/mg/min) | Relative Activity (%) | |
|---|---|---|
| Actinidia arguta | 16.97 | 100.0 |
| Actinidia deliciosa | 7.70 | 45.4 |
| Actinidia chinensis | 0.025 | 0.015 |
| Actinidia polygama | 0.83 | 4.89 |
| Step | Total Protein (mg) | Total Activity (units) | Specific Activity (μmoles/mg/min) | Purification (-fold) | Yield (%) |
|---|---|---|---|---|---|
| Dialysis | 70.35 | 1980 | 28.15 | 1 | 100 |
| UNOsphere Q | 20.59 | 1228.8 | 59.68 | 2.12 | 62.1 |
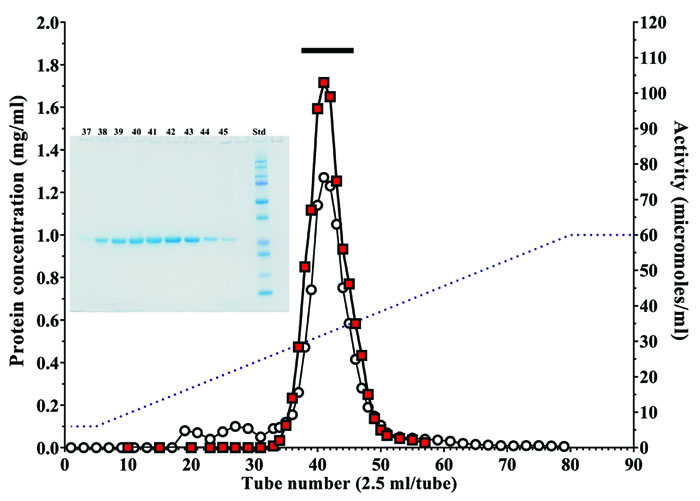
Fractions containing cysteine protease activity were collected. The bar indicates the fractions that were pooled. Protein concentration (mg/ml) (○), cysteine protease activity (■) and sodium chloride gradient (--). Insert: SDS-PAGE of fractions containing the enzyme activity. 3 μl of each aliquot (fraction numbers 37 to 45) was subjected to SDS-PAGE.
3.4. Biochemical Properties of the Purified Enzyme
3.4.1. Molecular Weight
Cysteine protease from A. arguta fruit juice was purified to homogeneity, and the molecular weight of the enzyme was calculated to be approximately 25,500 on SDS-PAGE (Fig. 3). The value of its molecular weight was close to those of enzymes from other plants including actinidin (Mr 23,000 ~ 24,000) [18, 24, 25], papain (Mr 23,400) [26] and ficain (ficin) (Mr 23,400) [27].
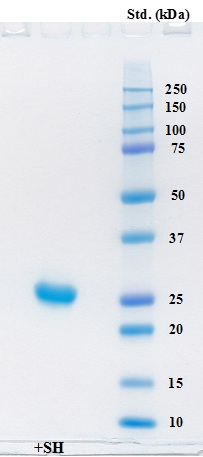
Electrophoresis of the purified enzyme was carried out on a Mini-protean TGX precast gel (any kD) in the presence of SDS and the gel was stained with Coomassie Brilliant Blue G-250. Lane 1 contained 5 μg of the purified protein in the presence of β-ME. Precision Plus protein standards (Bio-Rad) were also used for estimation of molecular weight.
3.4.2. Substrate Specificity
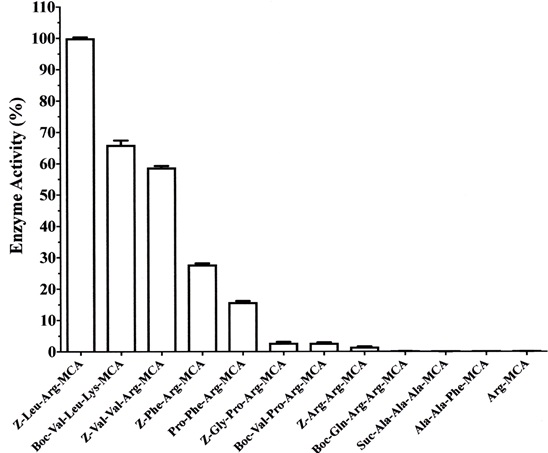
As shown in Fig. (4), cysteine protease from Actinidia arguta exhibited high activity toward the synthetic substrate Z-Leu-Arg-MCA and moderate activity toward Z-Val-Leu-Lys-MCA, Z-Val-Val-Arg-MCA, Z-Phe-Arg-MCA and Pro-Phe-Arg-MCA. Furthermore, the enzyme very weakly hydrolyzed the substrates Z-Gly-Pro-Arg-MCA, Boc-Val-Pro-Arg-MCA and Z-Arg-Arg-MCA. It did not hydrolyze substrates for cathepsin H (Arg-MCA), elastase (Suc-Ala-Ala-Ala-MCA) and tripeptidyl peptidase II (Ala-Ala-Phe-MCA).
Our results indicated that the enzyme preferentially cleaves the peptide bond between P1 (Arg or Lys) and P’1 positions in the presence of a hydrophobic amino acid such as Leu, Val and Phe at the P2 position.
3.4.3. Optimal pH, pH Stability, Optimal Temperature and Thermo-stability
Cysteine protease activity was assayed in the pH range of 3.0 to 8.0 using citrate and sodium-phosphate buffers. The activity of cysteine protease toward the substrate Z-Leu-Arg-MCA was optimal at pH 6.5 - 7.5 when sodium phosphate buffer was used (Fig. 5A).
To examine the effect of pH on stability, the enzyme was incubated at various pH values (GTA buffer: 3.5 to 10.0) for 24 hr at 4°C. The enzyme was stable over a wide pH range from 4.0 to 8.5 (Fig. 5B).
The temperature for the optimal hydrolysis of the above substrate was approximately 50°C (Fig. 5C).
The enzyme activity was thermo-stable up to 45°C for 10 min of incubation (Fig. 5D).
In 20 mM sodium phosphate buffer, pH 7.0, the enzyme was stable for at least 7 days at 4°C and three months at -30°C.
3.4.4. Kinetic Parameters
The Km, Vmax, Kcat and Kcat/Km values of the purified enzyme for four substrates at optimal pH (pH 7.0) were determined (Table 2). The Km, Vmax, Kcat and Kcat/Km values for Z-Leu-Arg-MCA, which was most preferentially cleaved by the enzyme, were 100 μM, 63.8 μmoles/mg/min, 27.12 sec-1 and 0.2712 sec-1μM-1, respectively. The relative order of Kcat was Z-Leu-Arg-MCA > Boc-Val-Leu-Lys-MCA ≥ Z-Val-Val-Arg-MCA > Z-Phe-Arg-MCA and the relative order of Kcat/Km was Z-Leu-Arg-MCA > Boc-Val-Leu-Lys-MCA > Z-Val-Val-Arg-MCA = Z-Phe-Arg-MCA. These results indicated that Z-Leu-Arg-MCA is the best substrate among the four substrates and coincided with the results shown in Fig. (4).
3.4.5. Inhibition of Enzyme Activity by Standard Protease Inhibitors
Various protease inhibitors against cysteine protease from A. arguta were examined (Fig. 6). Cysteine protease activity was strongly inhibited by antipain, leupeptin, E-64 and E-64c, and Z-LLL-H (aldehyde) and was moderately inhibited by pepstatin A. However, the enzyme activity was not inhibited by EDTA, AEBSF, CA-074 and leuhistin. Accordingly, the enzyme from A. arguta was confirmed to belong to the cysteine protease family.
Each assay was carried out at 37°C for 10 min in 50 mM sodium phosphate buffer, pH 7.0, in the presence of the purified enzyme (0.5 μg) and 100 μM of a substrate. Each value is the mean ± SE from triplicate experiments.
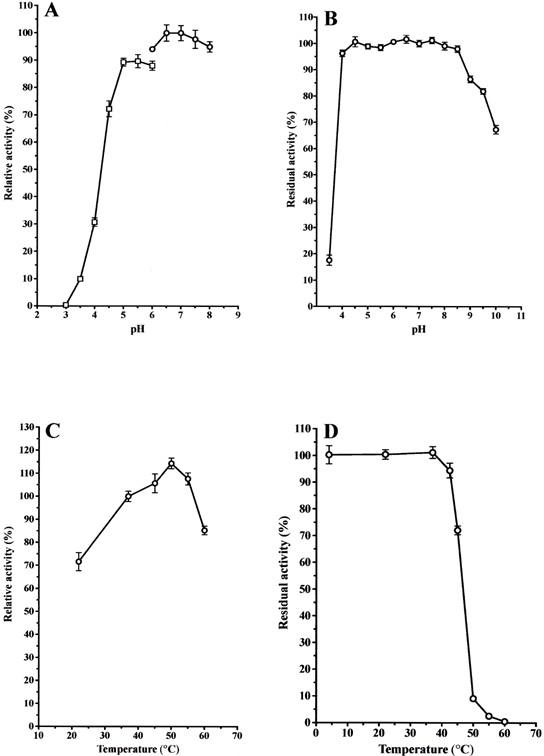
(A): The purified enzyme (0.5 μg) was incubated in 50 mM citrate buffer and sodium phosphate buffer at various pHs (3.0 ~ 8.0) for 10 min at 37 °C. The activity for Z-Leu-Arg-MCA was determined, and the activity obtained at pH 7.0 was useded as the standard (100%). Each value is the mean ± SE from triplicate experiments. (○): sodium phosphate buffer; (□): sodium citrate buffer. (B): The purified enzyme (0.5 μg) was stored in 50 mM GTA buffer at various pHs (3.5 ~ 10.0) for 24 hr at 4°C, and then the activity for Z-Leu-Arg-MCA was determined. The activity obtained at pH 7.0 was used as the standard (100%). Each value is the mean ± SE from triplicate experiments. (C): The purified enzyme (0.5 μg) was incubated in 50 mM sodium phosphate buffer, pH 7.0, at various temperatures (22 ~ 60°C) for 10 min. The activity for Z-Leu-Arg-MCA was determined, and the activity obtained at 37 °C was used as the standard (100%). Each value is the mean ± SE from triplicate experiments. (D): The purified enzyme (0.5 μg) was pre-incubated in 50 mM sodium phosphate buffer, pH 7.0, at various temperatures (22 ~ 60°C) for 10 min and then incubated for an additional 10 min after an addition of the substrate. The activity obtained at 22°C was used as the standard (100%). Each value is the mean ± SE from triplicate experiments.
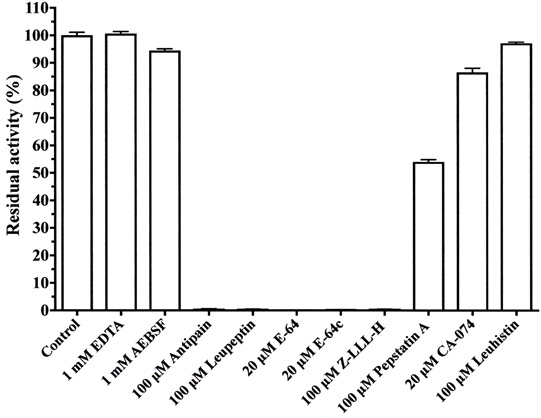
The purified enzyme (0.5 μg) was pre-incubated with each inhibitor for 10 min at room temperature in 50 mM sodium phosphate buffer, pH 7.0, and assays were started by adding 100 μM Z-Leu-Arg-MCA. The reaction mixtures were incubated for an additional 10 min at 37°C. Each value is the mean ± SE from triplicate experiments.
| Substrate | Km (μM) | Vmax (μmoles/mg/min) | Kcat (sec-1) | Kcat/Km (sec-1μM-1) |
|---|---|---|---|---|
| Z-Leu-Arg-MCA | 100 | 63.8 | 27.26 | 0.2726 |
| Z-Val-Val-Arg-MCA | 263 | 41.4 | 17.69 | 0.0672 |
| Boc-Val-Leu-Lys-MCA | 91 | 41.5 | 17.74 | 0.1949 |
| Z-Phe-Arg-MCA | 222 | 34.9 | 14.91 | 0.0671 |
| Inhibitor | IC50 (nM) |
|---|---|
| E-64 | 4.5 |
| E-64c | 11.5 |
| CA-074 | 26,500.0 |
| Leupeptin | 55.0 |
| Antipain | 93.0 |
| Z-LLL-H | 9.0 |
| Pepstatin A | 160.0 |
| Kinin-free LMW kininogen | 4.6 |
| Cystatin C | 30.5 |

The N-terminal amino acid sequence of cysteine protease from Actinidia arguta is aligned against the sequences of cysteine proteases from Actinidia deliciosa (GenBank accession no. ABQ1090), Actinidia eriantha (ABQ 10191), Carica Papaya (AAB 02650) and Ananas comsus (fruit (BAA 21929) and stem (CAA 08860.1)). Non-conserved amino acid residues among the cysteine proteases of those fruits are shown in filled boxes.
The 50% inhibition (IC50) values of inhibitory activity of various inhibitors toward cysteine protease purified from A. arguta were determined with a fixed substrate concentration of 100 μM but with various inhibitor concentrations (Table 3).
The relative order of IC50 values of synthetic inhibitors was E-64 > Z-LLL-H > E64c > leupetin > antipain. Furthermore, two proteinase inhibitors, kinin-free LMW (low molecular weight) kininogen [28] and cystatin C [29], strongly inhibited the activity of the enzyme (IC50 = 3.9 and 32.0 nM, respectively) [30]. LMW kininogen is well known to have three homologous copies of cystatin C, and among the three domains, domain 2 and 3 strongly inhibit the activity of cysteine protease. Accordingly, it is thought that the difference in the IC50 values of the two inhibitors depends on their number of inhibitor domains [31-33] (Table 4).
3.4.6. Comparison of the Amino Acid Sequences of Cysteine Proteases Between A. arguta and Other Plants
As shown in Fig. (7), the amino acid sequence of the first 21 residues of cysteine protease purified from A. arguta was Val1-Leu-Pro-Asp-Tyr5-Val-Asp-Trp-Arg-Ser10-Ala-Gly-Ala-Val-Val15-Asp-Ile-Lys-Ser-Qln20-Gly. However, both valine1 and leucine2 were detected with has been a 1:1 ratio as the N-terminal amino acid (figure not shown). On the other hand, it has been reported that the N-terminal amino acid of actinidin from Actinidia delisiosa and that of papain from C. papaya started at the 2nd position (Fig. 7) [24, 34]. It is thought that these proteases are synthesized as precursor forms and that their N-terminal regions are cleaved by the processing enzymes. However, the processing enzymes have not yet been identified. There is a possibility that in the case of cysteine protease from A. arguta fruit juice, the processing enzyme may not work sufficiently during its processing.
The primary function of endogenous cysteine proteases in plants is thought to be protection against arthropod pests, plant-parasitic nematodes and herbivorous insects invading the plants [35, 36]. Endogenous cysteine proteases are also thought to be involved in many physiologically important processes such as disease resistance, programmed cell death, germination and senescence [37-41].
On the other hand, cysteine proteases such as papain, bromelin, ficin, actinidin and zingibain from some fruits have been extensively and effectively utilized for production of tender meat [12-16, 19, 42, 43], although their characteristics including optimal pH, optimal temperature and enzymatic kinetics for the production of tender meat have not been determined.
Montoya et al. [44, 45] reported that dietary actinidin from kiwifruit (A. deliciosa cv. Hayward) increased gastric digestion and gastric emptying rate of several dietary proteins in growing rats [44] and meat proteins in growing pigs [45]. Accordingly, cysteine protease from A. arguta fruit juice is also expected to have the same nutritional effects.
On the other hand, Cavic et al. [46] and Grozdanovic et al. [47] reported that active actinidin is resistant to Simulated Gastric Fluid (SGF) and Simulated Intestinal Fluid (SIF) and can influence/increase the uptake of complete actinidin molecules and other barely degraded proteins by breaking down occludin (a key epithelial tight junction transmembrane protein) in the tight junction network. These findings indicate that food allergens with intrinsic proteolytic activity can endanger the gut epithelial barrier, thus contributing to the sensitization process in food allergy pathogenesis.
Functional and processed foods that utilize the distinctive feature of cysteine protease from A. arguta fruit juice to promote protein digestion and absorption should be developed, and foods such as A. arguta and kiwifruit (A. deliciosa cv. Hayward) are also expected to contribute to the maintenance and improvement of human health through their proteolytic functions.
LIST OF ABBREVIATIONS
| AEBSF | = 4-(2-aminoethyl)-benzenesulfonyl fluoride |
| AMC | = 7-amino-4-methylcoumarin |
| Boc- | = t-butyloxycarbonyl- |
| β-ME | = β-mercaptoethanol |
| E-64 | = N-[N-(L-3-trans-carbxyran-2-carbonyl)-L-leucyl]agmatine |
| EDTA | = ethylendiaminetetraacetic acid |
| LMW | = low molecular weight |
| MCA | = 4-methyl-coumaryl-7-amide |
| PAGE | = polyacrylamide gel electrophoresis |
| PMSF | = phenylmethylsulfonyl fluoride |
| Z- | = benzyloxycarbonyl- |
| SDS | = sodium dodecyl sulfate |
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This work was supported in part by grants from the Tenshi College Research Foundation (to I. Ohkubo).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


