All published articles of this journal are available on ScienceDirect.
In Vitro Analysis of Protein:Protein Interactions in the Human Cancer-Pertinent rp.eL42-p53-Mdm2 Pathway
Abstract
Introduction:
We have recently demonstrated that the eukaryote-specific large subunit ribosomal protein (rp) eL42 assists catalysis of peptide bond formation at the peptidyl transferase center of 80S ribosomes in eukaryotic cells. Recently, several ribosomal proteins were shown to have extraribosomal functions independent of protein biosynthesis. Such functions include regulation of apoptosis, cell cycle arrest, cell proliferation, neoplastic transformation, cell migration and invasion, and tumorigenesis through both Mdm2-p53-dependent and p53-independent mechanisms. Our objective is to demonstrate that overexpression of eL42 in tumor may incapacitate cell anti-tumor mechanism through interaction with the tumor suppressor protein p53 and its partner Mdm2.
Methods:
Co-immunoprecipitation technique and the binding assays on Biacore were used to probe interactions between recombinant eL42, p53 and Mdm2 proteins in a so-called rp-p53-Mdm2 axis.
Results:
We demonstrate that the ribosomal protein eL42, the tumor suppressor protein p53 and the ubiquitin E3 ligase Mdm2 interact with each other in a ternary rp.eL42:p53:Mdm2 complex. Precisely, the interaction between eL42 and p53 is characterized by a strong binding affinity (KD value in the nanomolar range) that is likely to trigger the sequestration of p53 and the inhibition of its tumor suppressor activity. Furthermore, the p53:Mdm2 and eL42:Mdm2 complexes exhibit comparable binding affinities in the micromolar range compatible with Mdm2 being the enzyme which ubiquitinates both the p53 and eL42 substrates. Interestingly, pyridoxal 5'-phosphate (PLP), one of the active forms of vitamin B6, binds to eL42 and significantly inhibits the interaction between eL42 and p53, in accordance with the observation that vitamin B6 is associated with reduced risk of cancer.
Conclusion:
Our study emphasized one more major mechanism of p53 downregulation involving its sequestration by eL42 upon the overexpression of this ribosomal protein. The mechanism described in the present report complemented the well-known p53 downregulation triggered by proteasomal degradation mediated through its ubiquitination by Mdm2.
1. INTRODUCTION
It is widely accepted that cancer is a disease characterized by anarchic cell proliferation that can be stopped by tumor suppressor proteins [1]. The tumor suppressor protein p53 is well known as the “guardian of the genome” as it is essential for genomic stability [1]. P53 importance is highlighted by the fact that it is mutated in at least half of all human cancers, while it is functionally inactivated in much of the remaining 50% of cancers through signaling pathways [2-6]. The two major essential negative regulators of p53 are Mdm2 (Mouse Double Minute 2 also known as Hdm2) and its closely related homolog Mdm4 also known as MdmX or HdmX [2-6]. As a consequence, the Mdm2-p53 axis is an important pathway frequently deregulated in cancer. In fact, Mdm2 is an ubiquitin E3 ligase which plays a key role in inhibiting p53 under both physiological and stress conditions. Mdm2 inhibition of p53 consists in ubiquitinating p53 and targeting it for proteasomal degradation [7, 8]. Finally, several ribosomal proteins (rps) were previously shown to be involved in the regulation of the Mdm2-p53 axis through specific interactions with Mdm2 or p53 following ribosomal stress [9-12]. One of these rps is the eukaryote-specific large subunit ribosomal protein eL42 (formerly L42A or L42AB in yeast, L36a or L36a-like in human, or L44e in archaea). The ribosomal protein eL42 (rp eL42) presents the following characteristics: (i) it was recently shown to directly and actively contribute to the activity of 80S ribosomes at the elongation step of translation [13], thus suggesting that this rp might control the rate of protein biosynthesis in health and in disease; (ii) it was found to be overexpressed in human hepatocellular carcinoma as well as in several human tumor cell-lines, suggesting that its extra-ribosomal role might be related to tumor cell proliferation [14]; (iii) in the crystallographic structure of S. cerevisiae 80S ribosomes or of the 50S subunit of Haloarcula marismortui, most of the anticancer drugs were shown to target the eL42 protein [15-19]. These observations point out a connection between protein synthesis on the ribosome and cancer cells growth. In fact, increasing the rate of protein synthesis would be favourable to the increase in cancer cell size and to their subsequent division and growth. In this view, it is interesting to note that rp eL42 is a target for cycloheximide, a strong inhibitor of ribosomal protein biosynthesis in eukaryotic cells that has proven to be a potent anti-tumor drug [19]. Altogether, these observations suggested that eL42 might be a candidate target for anticancer therapy, while the interactions between p53, Mdm2 and eL42 might be a crucial event in tumorigenesis. In the present report, we probed the interactions between p53, Mdm2 and eL42 by performing protein-protein binding assays on Biacore. We demonstrate that the interactions between these proteins take place with strong binding affinities, in accordance with the previous reports on the downregulation of the tumor suppressor p53 by Mdm2, on one hand, or by the ribosomal protein eL42 alone or in combination with Mdm2, on the other hand. Finally, we demomonstrate that pyridoxal 5'-phosphate (PLP), a small-molecule inhibitor representing one of the active forms of vitamin B6, specifically targets eL42 and significantly inhibits the eL42-p53 interaction.
2. MATERIALS AND METHODS
2.1. Materials
For expression and purification of the recombinant proteins, the E. coli Solu BL21 cells (AMS Biotechnology) were used. Penta.His biotin conjugate and Ni-NTA superflow columns were from Qiagen. Hi-load 16/60 Superdex 75 and His graviTrap columns were from GE healthcare. Amicon ultrafiltration apparatus (Millipore membrane PM10) was purchased from Thermo Fischer Scientific (France). Pwo DNA polymerase high fidelity PCR amplification was from Roche Diagnostics. Carbenicillin, isopropyl-ß-D-thiogalacto-pyranoside, PBS, tris-HCl, NaCl, EDTA, MgCl2, glycerol, ß-mercapto-ethanol, SDS, imidazole, lysozyme, tween-20, DNAase and other chemicals were from Sigma-Aldrich (France). For co-immunoprecipitation and western blotting experiments the following primary antibodies were used: mouse anti-p53 (Santa Cruz), mouse anti-RPL36a (Santa Cruz) and mouse anti-MDM2 (Santa Cruz). complete ™, Mini, EDTA-free Protease Inhibitor Cocktail was from Sigma Aldrich. nProtein A Sepharose 4 Fast Flow was from GE Healthcare. Tris-Base, glycine, sodium dodecyl sulfate, bis-acrylamide and nitrocellulose membrane and MagicMark™ XP Western Protein Standard were from Thermo Fischer Scientific (France). The HRP-conjugated secondary antibodies (mouse IgG, rabbit IgG) were purchased from GE Healthcare. For protein detection on western blots, the enhanced chemiluminescence substrate reagent was purchased from Perkin Elmer (USA).
2.2. Methods
2.2.1. Expression and Purification of the Human Recombinant eL42 Protein
His-Tagged eL42 was expressed in Solu BL21 E. coli cells harboring the plasmid pColdI-eL42. A fresh overnight culture was used to inoculate 5 liters of LB with 50 μg/ml carbenicillin and grown with shaking at 37°C to 0.5 OD600, after which induction with 0.4 mM isopropyl-ß-D-thiogalacto-pyranoside took place at 15°C with incubation up to 30 hours. After harvesting, bacteria (8.5g) were washed in PBS, sonicated 15 times for 10 s at 4°C in 40 ml buffer A (25 mM Tris-HCl pH7.5, 0.5 M NaCl, 10% glycerol, 10 mM Imidazole, 1 mM ß-ME) containing 0.05% Tween 20, 1 mM MgCl2, 0.5 mg/ml lysosyme, 100 μg/ml DNase, one tablet of complete mini EDTA free protease inhibitor cocktail, and centrifuged (100,000 g for 30 minutes). Western blot analysis on supernatant and pellet using penta.His biotin conjugate revealed that eL42 was nearly insoluble. Consequently, the pellet was suspended in 30 ml buffer A containing 8 M urea and sonicated 30 times for 6 s. The supernatant obtained by centrifugation (100,000 g for 30 minutes) was applied two times to Ni-NTA superflow column (2 ml), and washed with 20 ml buffer A containing 8 M urea and 25 mM Imidazole. The protein was then eluted with 10 ml buffer A containing 8 M urea and 0.4 M imidazole. This fraction was concentrated to 1.5 ml in an Amicon ultrafiltration apparatus (Millipore membrane PM10) and applied to Hi-load 16/60 Superdex 75 column equilibrated in buffer B (50 mM Tris-HCl pH7.5, 0.35 M NaCl, 7 mM ß-ME). Fractions eluted containing purified eL42 (5 mg), free of urea were concentrated by ultrafiltration and dialyzed against buffer B with 50% glycerol and stored at -25°C.
2.2.2. Cloning, Expression and Purification of Human p53
Purification of p53•GST. Full length p53 was expressed in Solu BL21 E. coli cells (AMS Biotechnology) harboring the plasmid pGEXp53FL as fusion with glutathione S-transferase. The transformed E. coli strain was grown at 37°C in 2 liters of LB rich medium containing 50 µg.mL-1 ampicillin to 0.5 A600, after which induction with 0.4 mM isopropyl-ß-D-thiogalacto-pyranoside took place at 20°C with incubation up to 20 hours. After harvest, the cells were washed in PBS, sonicated 15 times for 10 s at 4°C in 40 ml buffer A (50 mM Tris-HCl pH7.5, 150 mM NaCl, 7 mM ß-ME, 10% glycerol, containing 0.5 mg.ml-1 lysosyme, 20 µg.ml-1 DNase, one tablet of complete mini EDTA free protease inhibitor cocktail (Roche Diagnostics), and centrifuged (100.000 g for 30 minutes). The extract supernatant was applied on a 3 ml glutathione sepharose 4 fast flow column (GE Healthcare) equilibrated with buffer A. The colum was washed with 5 volumes buffer A, then with 5 volumes buffer B (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 7 mM ß-ME, 10% glycerol. p53•GST was eluted at least 90% pure with 3 volumes of buffer B containing 10 mM reduced glutathione. This fraction was concentrated by ultrafiltration, dialysed against buffer C (50 mM Tris-HCl pH 7.5, 0.3 M NaCl, 1 mM dithiothreitol, 50% glycerol) and stored at -80°C.
Purification of His-Tagged p53. His tagged p53 DNA was obtained after Pwo DNA polymerase high fidelity PCR amplification, using pGEXp53FL as template and the 5’ primer CACCATGGAGGAGCCGCAGTCAGATCCTAGC, and 3’ primer TCAGTCTGAGTCAGGCCCCTTCTGTCTTG, followed by cloning into pET151/DTOPO vector according to the TOPO cloning procedure (Invitrogen). This vector allows expression of recombinant protein with an N-terminal containing the V5 epitope, a 6XHis tag and a TEV protease cleavage site. Accuracy of the cloning was analyzed by sequencing. BL21(DE3) bacteria cells were transformed with pet151/D-TOPO/p53 plasmid. Bacteria were grown at 37°C in 2 liters of LB rich medium containing 50 µg. mL-1 carbenicillin to 0.5 A600, after which induction with 0.1 mM isopropyl-ß-D-thiogalacto-pyranoside took place at 20°C with incubation overnight. After harvest, the cells were washed in PBS, sonicated 15 times for 10 s at 4°C in 40 ml buffer D (25 mM Tris-HCl pH 7.5, 0.5 M NaCl, 1 mM ß-ME, 10 mM Imidazole, 0.05% Tween 20, 10% glycerol, 1 mM MgCl2 containing 0.5 mg.ml-1 lysosyme, 100 µg.ml-1 DNase, one tablet of mini EDTA free protease inhibitor cocktail (Roche Diagnostics), and centrifuged (100, 000 g for 30 minutes). Western blot analysis on supernatant and pellet using penta.His biotin conjugate revealed that His-Tagged p53 was totaly insoluble, partially soluble in a variety of detergents such as 1% Triton, 40 mM n-octyl glucoside, 0.5% n-lauryl sarcosyl, 13 mM Chaps, 50mM hecameg, 1% Tween 20 but completely soluble in urea 8M. Consequently, the pellet was suspended in 30 ml buffer D containing 8M urea and sonicated 12 times for 6 s at 4°C. The supernatant obtained by centrifugation (100, 000 g for 30 minutes) was very partially retained to Ni-NTA superflow column (2 ml), nevertheless, the 8M urea containing supernatant showed His-Tagged p53 almost pure. This supernatant was concentrated in an Amicon ultrafiltration apparatus (Millipore membrane PM10) and applied to Hi-load 16/60 Superdex 75 column (Äkta purifier system) equilibrated in buffer E (50 mM Tris-HCl pH7.5, 0.35 M NaCl, 7 mM ß-ME, 10% glycerol). Eluted fractions containing purified His-Tagged p53 free of urea were concentrated by ultrafiltration, aliquoted at a concentration of 5 mg.ml-1 to avoid precipitation and stored at -80°C.
2.2.3. Cloning, Expression and Purification of the Human His-tagged Mdm2 [1-350]
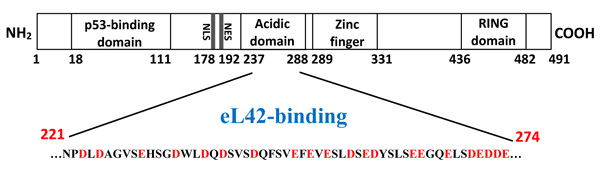
First attempts to purify the full length Mdm2 revealed that this protein was nearly insoluble. Failure to obtain a full length Mdm2 protein in a native soluble form prompted us to engineer a fragment [1-350] as follows: (i) His tagged Mdm2 [1-350] fragment DNA was obtained after Pwo DNA polymerase high fidelity PCR amplification, from pGEXMdm2 as template and the 5’ primer CACCATGTGCAATACCAACATGTCTGTACCTAC, and 3’ primer CTATGAGTTTTCCAGTTTGGCTTTCTCAGAGAT ; (ii) the amplified fragment was purified and cloned into pET151/D-TOPO following the TOPO cloning procedure. Accuracy of the cloning was analyzed by sequencing; (iii) BL21(DE3) bacteria were transformed with pet151/D-TOPO/Mdm2[1-350] plasmid. Bacteria were grown at 37°C in 3 liters of LB rich medium containing 50 µg.mL-1 carbenicillin to 0.5 A600, after which induction with 0.1 mM isopropyl-ß-D-thiogalacto-pyranoside took place at 23°C with incubation overnight ; (iv) after harvest, the cells were washed in PBS, sonicated 15 times for 10 s at 4°C in 40 ml buffer D containing 0.5 mg.ml-1 lysosyme, 100 µg.ml-1 DNase, one tablet of complete mini EDTA free protease inhibitor cocktail ; (v) the supernatant was applied two times on 1 ml His graviTrap column equilibrated in buffer D. After washing with 20 ml buffer D containing 20 mM imidazol, followed by step elution in buffer D containing, 50, 100, 250, 400 mM imidazole respectively, Mdm2 (1-350) was eluted almost pure in 10 ml of the fraction containing 250 mM imidazol as two bands revealed at a molecular weight of about 50 kDa on SDS PAGE electrophoresis ; (vi) this fraction was concentrated in an Amicon ultrafiltration apparatus (Millipore membrane PM10), dialyzed against 50 mM Tris-HCl pH 8.0, 60 mM KCl, 7 mM ß-ME (buffer F), loaded on Mono Q-HR 5/5 using Äkta purifier system and purified in a linear 0.1-0.7 M KCl in buffer F. The pure eluted fractions were concentrated, dialyzed against buffer F containing 50% glycerol, 1 mM dithiothreitol, and stored at -30°C. As shown in Fig. (1), the presence of the binding sites for proteins eL42 and p53 on Mdm2 [1-350] indicates that this fragment is suitable for the study of protein:protein interactions in the cancer-pertinent rp.eL42-p53-Mdm2 pathway.
2.2.4. The In Vitro Co-immunoprecipitation (Co-IP) Technique
Co-immunoprecipitation is the main method of studying protein:protein interactions since it is possible to use endogenous, tagged and/or overexpressed proteins. Whether the protein of interest is purified or present in a cell lysate, it can be immunoprecipitated with a specific antibody linked to protein A beads coupled to sepharose. Since the conditions are non-denaturing, all the proteins interacting with the protein of interest will be precipitated as well. These protein complexes are then denatured and the interaction partners are analyzed on denaturing gel (SDS-PAGE) followed by Western blotting. This method can detect both direct and indirect interactions. The analysis of this complex makes it possible to identify new binding partners and their binding affinities. The eL42 protein was incubated for 1h at 4°C with its specific antibody in an immunoprecipitation (IP) buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1% NP-40, 2% Triton X-100 and 2.5% Glycerol. After incubation, protein A-Sepharose beads previously washed 3 times with IP buffer were added to the reaction mixture followed by incubation for 1 hour at 4°C. Then, the p53 protein was added to this complex, followed by wheel incubation at 4°C overnight. The next day, the co-IP was stopped by centrifugation at 14.000 g for 10 min at 4°C, the supernatant (SN) which should contain putative p53 not associated with eL42, was recovered and stored for deposit. The pellets (P), which should contain eL42 and putative associated p53, were then washed 4 times for 10 min on a wheel at 4°C with the following washing solutions: washing 1 with (400 mM NaCl, 10 mM Tris-HCl pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.5% BSA); washing 2 with (400 mM NaCl, 10 mM Tris-HCl pH7.5, 1 mM EDTA, 1% Triton X-100); washing 3 with (150 mM NaCl, 10 mM Tris-HCl pH7.5, 1 mM EDTA, 1% Triton X-100) and washing 4 with (150 mM NaCl, 10 mM Tris-HCl pH7.5, 1 mM EDTA). Finally, the elution of the complex formed is carried out with the denaturation Laemmli buffer and heated at 95°C for 5 min. Equal fractions of proteins were analyzed by SDS-PAGE followed by immunodetection using either anti-p53 antibody (DO7) or anti-eL42 antibody to verify the interaction between the target proteins. The same technique was used to show the interactions between Mdm2 and eL42.
2.2.5. Poly(U)-Dependent Poly(Phe) Synthesis Activity of Human 80S Ribosomes in the Presence of Human Recombinant rp eL42
Poly(Phe) synthesis was determined as incorporation of L-[14C(U)]Phenylalanine into hot trichloroacetic acid-insoluble material as described in [20]. The reaction mixture (100 μl) contained 40 mM Tris-HCl pH 7.5, 7 mM MgCl2, 80 mM NH4Cl, 1 mM dithiothreitol, 1 mM ATP, 1 mM Phosphoenolpyruvate, 0.3 mM creatine phosphate, 0.5 mM GTP, 50 μg.ml-1 pyruvate kinase, 50 μg.ml-1 creatine kinase, 5 μM tRNA (first charged during a 30 min incubation at 30°C with a 2 fold excess of L-[14C(U)]Phenylalanine (5 GBq.mmol) and a saturating amount of partially purified phenylalanyl-tRNA synthetase, 3.5 μg poly(U) and 0.5 μM EF-1α, 0.15 μM EF-1β, 0.35 μM EF-2, and 14 pmol of human 80S ribosome. During incubation at 30°C, 30 μl aliquots were withdrawn at times indicated, spotted on glass fiber filters and hot trichoroacetic acid-insoluble radioactivity was determined. Effect of increasing concentrations of eL42 on the in vitro poly(U)-dependent poly(Phe) synthesis activity of human 80S ribosomes. The reaction was conducted as follows: (i) increasing concentrations of eL42 were preincubated for 10 min at 30°C with charged tRNAPhe (5 μM) in 30 μl of the aforementioned incubation mixture; (ii) the reaction was started with ribosomes and the activity was determined after 40 min incubation at 37°C. Another variant of activity determination consisted in adding eL42 to the incubation mixture of the poly(U)-dependent poly(Phe) synthesis reaction catalyzed by human 80S ribosomes when [14C]Phe incorporation has reached a plateau value within 40 min. Then, the activity was determined with (triangles) or without (circles) addition of eL42 (35 μM final concentration).
2.2.6. Surface Plasmon Resonance
The surface plasmon resonance (SPR) biosensor experiments were performed on a Biacore 3000 instrument (GE Healthcare) of the Platform of Molecular Interactions of the Institute of Biology Paris Seine (IBPS, Sorbonne University). All experiments were performed in triplicate, as described previously [21, 22]. The CM5 sensor chip was used. The His-tagged purified eL42 protein (called the ligand) was immobilized through primary amino groups to the carboxymethyl dextran matrix of a CM5 sensor chip. His-tagged eL42 (1 μM) diluted in immobilization buffer (10 mM sodium acetate, pH 5.5) was injected at a flow rate of 10 μl/min for 7 minutes in order to obtain 3900-5000 resonance units (RU) of covalently coupled eL42. The immobilization was followed by an injection of 70 μl of ethanolamine hydrochloride (1 M) pH 8.5, at a flow rate of 10 μl/min, to saturate the free activated sites of the matrix. A reference surface without protein was prepared using the same procedure. Buffer HBSEP (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) was used as the running buffer and for diluting all the injected proteins. The variations of SPR signal as a function of time (sensorgram) were obtained by passing various concentrations of the injected protein or small-molecule (called the analyte) over the ligand surface at a flow rate of 5 μl/min, with an association phase of 5 min and a dissociation phase of 8 min. The surface of the sensor was regenerated with an injection of 10 mM glycine hydrochloride (pH 2.0) at a rate of 30 μl/min for 30 seconds. When necessary, the regeneration was carried out by injection of 1M NaCl and 30 mM NaOH under the same conditions. Identical injections over blank surfaces executed in parallel (giving a value of 0 RU) were subtracted from all experiments. The kinetics were evaluated using the BIAevaluation software, version 4.1 (GE Healthcare). The data were processed by fitting the binding profiles to a 1:1 Langmuir interaction model. The quality of the fit was assessed by the statistical Chi2 value provided by the software (Chi2 values <10 were considered as acceptable). The fitting of each data set yielded rates for association (ka or kon) and dissociation (kd or koff), from which the dissociation constant KD was calculated (KD = koff/kon). The kon, koff and KD from 3 experiments were used to calculate the mean values of these variables.
3. RESULTS
3.1. Interactions between rp eL42 and the p53 or Mdm2 Proteins as Revealed by Co-immunoprecipitation and Western-Blotting
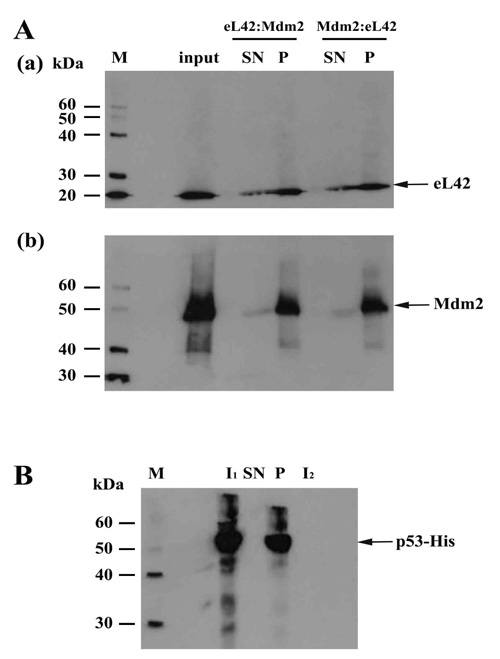
On one hand, the purified eL42 protein (0.032μg) was immunoprecipitated (IP) with anti-eL42 antibody and then incubated with purified p53-gst (0.027μg). In that case, a putative co-IP between eL42 and p53 is referred to as an eL42:p53 complex. On the other hand, p53-gst (0.027μg) was subjected to IP with anti-p53 antibody followed by incubation with purified protein eL42 (0.032μg). The putative corresponding complex between the two proteins is named p53:eL42 complex. For each complex, the pellet (P) and the supernatant (SN) fractions were both subjected to SDS-PAGE followed by western-blotting by using antibodies specific to each protein of the putative complex. As shown in Fig. (2a) (lane P), IP of eL42 was accompanied by the co-IP of p53, represented by a band of approximately 80 kDa (MW of p53 is 55 kDa and that of the GST-tag is 26 kDa). Fig. (2b) shows IP of p53 accompanied by the co-IP of eL42, represented by the 12 kDa band. These results suggest that the ribosomal protein eL42 interacts with the tumor suppressor protein p53 and reciprocally. In another set of experiments, the complex named eL42:Mdm2 was obtained with the purified eL42 protein (0.032 μg) subjected to IP with anti-eL42 antibody followed by incubation with 0.01 μg of Mdm2 [1-350] (Fig. 3A-(a)). Similarly, the Mdm2:eL42 complex was obtained by IP of 0.01 μg of the purified Mdm2 [1-350] protein with anti-Mdm2 antibody (SMP14) followed by incubation with eL42 (0.032 μg) (Fig. 3A-(b)). The samples were subjected to SDS-PAGE and the resulting gels to immuno-detection. Fig. 3A shows the immunoprecipitation of eL42 and the co-immunoprecipitation of Mdm2 and vice versa. These results suggest that the eL42 and Mdm2 proteins interact with each other. As a control, the purified Mdm2 [1-350] protein was immunoprecipitated with anti-Mdm2 antibody (SMP14) and then incubated with p53 (Fig. 3B). The interaction between the two proteins was detected by immunoblotting with anti-p53 (DO7) antibodies diluted to 1/500.000. These results confirm that the p53 and Mdm2 proteins interact with each other (Fig. 3B), as previously reported [23-25], and at the same time indicate that the IP/co-IP technique is a useful tool to demonstrate protein:protein interactions.
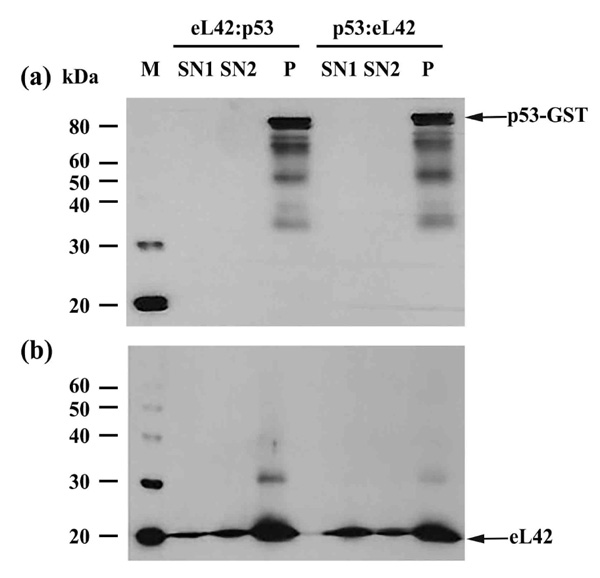
Immunoprecipitation of eL42 followed by incubation with p53, and vice versa, with (a): immunoblotting by means of anti-p53 (DO7) antibodies diluted 1/100.000; (b): detection of the interaction between eL42 and p53 with the anti-eL42 antibodies diluted 1/10.000. M: protein molecular mass standards; SN1: the supernatant after stopping the co-immunoprecipitation reaction; SN2: the first supernatant after the first wash; P: the pellet of immunocomplex containing the antibodies, the protein of interest and the antigen-associated proteins.
3.2. Interactions between the Human rp eL42 and the p53 or Mdm2 Proteins In Vitro
Surface plasmon resonance (SPR) analyses with a Biacore biosensor were carried out to determine the binding affinities of eL42 to p53 or Mdm2. All the proteins used in the Biacore assays were tagged with histidine. Therefore, they were immobilized on a CM5 sensor chip instead of a NTA sensor, in order to avoid interferences that could arise from the interaction with the sensor of the His-tag of both the analyte and the ligand. Fig. 4A shows a kinetic measurement of the interaction between human eL42 and p53. The corresponding kinetic and affinity constants, as deduced from Fig. 4A are: association (ka or kon) and dissociation (kd or koff) rates of 7.18×103 M-1. s-1 and 8.78×10-5 s-1, respectively, resulting in a binding affinity (dissociation constant KD) of 1.22 x 10-8 M. This KD value would reflect a good binding affinity between eL42 and p53. Another approach which allows an easy assessment of the strength of the eL42-p53 interaction is the determination of the half maximal effective concentration (EC50) of p53 that directs half-maximal binding of the tumor suppressor onto the ribosomal protein. As shown in Fig. (4B), the EC50 of the eL42-p53 interaction is 2.2 x 10-9 M. For the study of the eL42:Mdm2 interaction, a Mdm2 [1-350] fragment was engineered, as described in Materials and Methods. It contained all the functionally important domains of the protein, except the Mdm4 binding C-terminal domain (Fig. 1). The presence on this fragment of the binding sites for proteins eL42 and p53 indicates that it is suitable for the study of protein:protein interactions in the cancer-pertinent rp.eL42-p53-Mdm2 pathway. Fig. (4C) shows a kinetic measurement of the interaction between human eL42 and Mdm2 [1-350] where the kinetic and affinity constants are: association (ka or kon) and dissociation (kd or koff) rates of 180 M-1. s-1 and 1.28 × 10-3 s-1, respectively, resulting in a binding affinity (KD) of 7.09 × 10-6 M. Even though this KD value did not reflect a quite good binding affinity between eL42 and Mdm2, the results on Fig. (4C) would confirm that the eL42 and Mdm2 proteins interact with each other, as demonstrated with the IP/co-IP technique. Control experiments to assess the specificity of the protein:protein interactions studied in the present report consisted in analyzing (i) the binding on eL42 of anti eL42 antibodies (Fig. 4D) and (ii) the interactions between the eL42 protein and another RNA-binding protein that does not interact with the ribosome. In the latter case, we have used formyl-methionine-tRNA synthetase (FMTS), the enzyme which is in charge of transferring a formyl group onto the amino group of methionine esterified to the CCA-end of tRNAfMet. As shown in Figs. (4D and E), FMTS, even at a high concentration (500 nM) is not capable of binding to eL42, while anti eL42 antibodies were shown to specifically recognize eL42, as expected. Note that the highest concentration of p53 or Mdm2 used in the binding assays was only 50 nM (Fig. 4D and 4E).
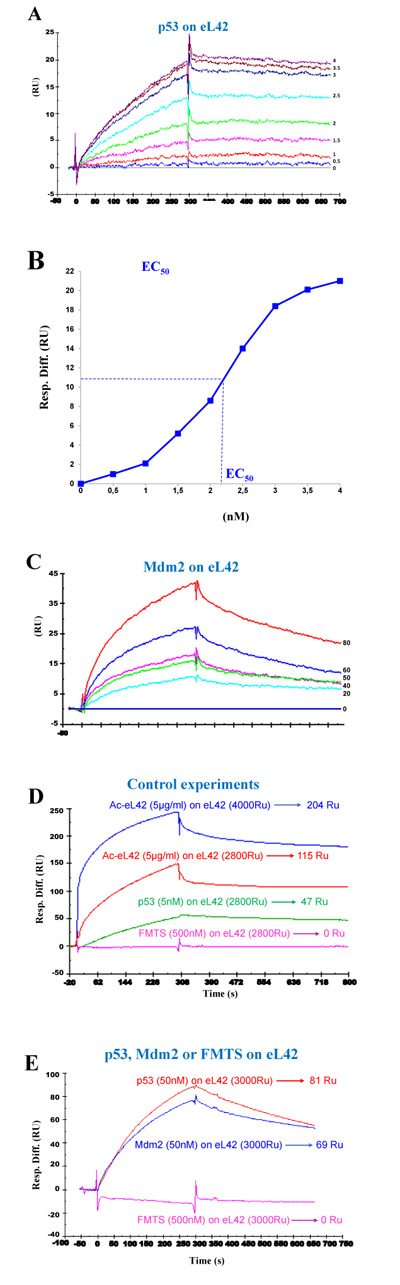
The His-tagged recombinant human eL42 was immobilized on the surface of CM5 sensor chip at low density resonance units (RU). (A): kinetics of interaction between eL42 and p53 at the concentrations of 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5 and 4 nM. (B): the half maximal effective concentration (EC50) of p53 corresponding to half-maximal saturation of eL42 was estimated to be 2.2 x 10-9 M. (C): kinetics of interaction between eL42 and Mdm2 at the concentrations of 0, 20, 40, 50, 60 and 80 nM. (D): specific binding of p53 to eL42 versus negative control (FMTS 500 nM) and specific recognition by the anti-eL42 antibodies on immobilized eL42. (E): demonstration of specific binding of p53 (50 nM) and Mdm2 (50 nM) on eL42 versus negative control (FMTS 500 nM).
3.3. Interaction between the Human p53 and Mdm2 Proteins In Vitro
The highly cancer-pertinent and well-defined p53-Mdm2 interaction has been studied during decades by several research groups. Determination of the binding affinities of p53 to Mdm2 (Fig. 5A) led to the following kinetic and affinity constants: association (ka or kon) and dissociation (kd or koff) rates of 429 M-1. s-1 and 9.5×10-4 s-1, respectively. The overall binding affinity (KD) was 2.21 x 10-6 M. Control experiments consisted in the absence of binding of FMTS on Mdm2 (Fig. 5B).
3.4. Pyridoxal 5'-phosphate Interferes with eL42-p53 Interactions
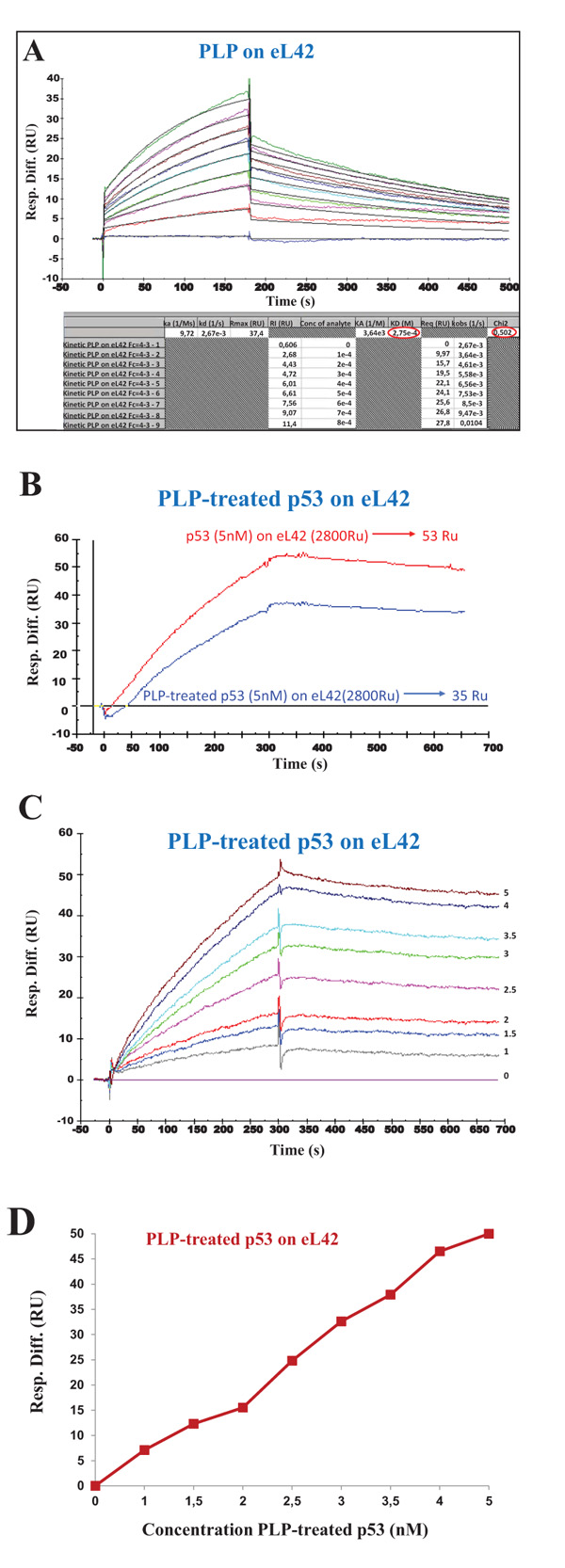
It is generally believed that small-molecule inhibitors might be capable of inhibiting protein:protein interactions in the p53:Mdm2 axis, in order to help develop new cancer therapeutics. One of these molecules is pyridoxal 5'-phosphate (PLP) which was previously shown to bind to the ribosomes and to inhibit their activity [26]. Interestingly, PLP is an active form of vitamin B6 which was proposed to exhibit anticancer properties [27-29]. These observations prompted us to check the effect of PLP on the interaction between eL42 and p53. However, a prerequisite for the study of the interaction between these proteins in the presence of PLP is to demonstrate that this small-molecule inhibitor is capable of binding on eL42 or p53 or both. In fact, if PLP doesn’t bind neither to eL42 nor to p53, it is most probable that it will not interfere with the interaction between these two proteins. In turn, if PLP binds to eL42 (12 kDa) which is much smaller than p53 (55 kDa), the region comprising the binding sites for PLP on eL42 would represent the exchange surface between these two partners, which would be much smaller than the total surface of the p53 protein. In that case, it is most probable that the interaction between eL42 and p53 would be affected in the presence of PLP. The potential interactions between PLP and eL42 were evaluated by investigating the kinetic parameters of the PLP-eL42 interaction by SPR. The sensorgrams shown in Fig. (6A) were obtained by injecting PLP at nine different concentrations (0, 100, 200, 300, 400, 500, 600, 700 and 800 µM) onto eL42 immobilized on CM5. Kinetic and affinity constants for the PLP-eL42 interaction, including association (ka or kon) and dissociation (kd or koff) rates and affinity constant (KD), as deduced from Fig. (6A) were, ka = 9.72 M-1.s-1 ; kd = 2.67 x 10-3 s-1 ; KD = 2.75 x 10-4 M. Then, the effect of PLP on the interaction between eL42 and p53 was checked as follows: (i) the His-tagged recombinant human eL42 was immobilized on the surface of CM5 sensor chip; (ii) the p53 samples referred to as PLP-treated p53 were all diluted in 1 mM PLP and immediately injected over the ligand surface; (iii) the concentrations of PLP-treated p53 run over the chip surface were: 0, 1, 1.5, 2, 2.5, 3, 3.5, 4 and 5 nM. It should be noted that, at 1 mM PLP, the rp eL42 was saturated by this small-molecule inhibitor as shown in Fig. (6A). Therefore, the variation of the SPR response versus p53 concentration would reflect only the binding of p53 on eL42. As shown in Fig. (6B), the extent of p53 binding on eL42 was found to be weaker in the presence of PLP than in its absence, suggesting that PLP interferes with the eL42-p53 interaction. The KD value for the p53:eL42 complex in the presence of PLP, as deduced from Fig. (6C) was 1.08 x 10-7 M which is tenfold higher than the KD value obtained in the absence of PLP (1.22 x 10-8 M). These results suggest that PLP is capable of significantly inhibiting the interaction between eL42 and p53. However, as shown in Fig. (6D), the EC50 of the interaction between eL42 and the PLP-treated p53 protein could not be obtained.
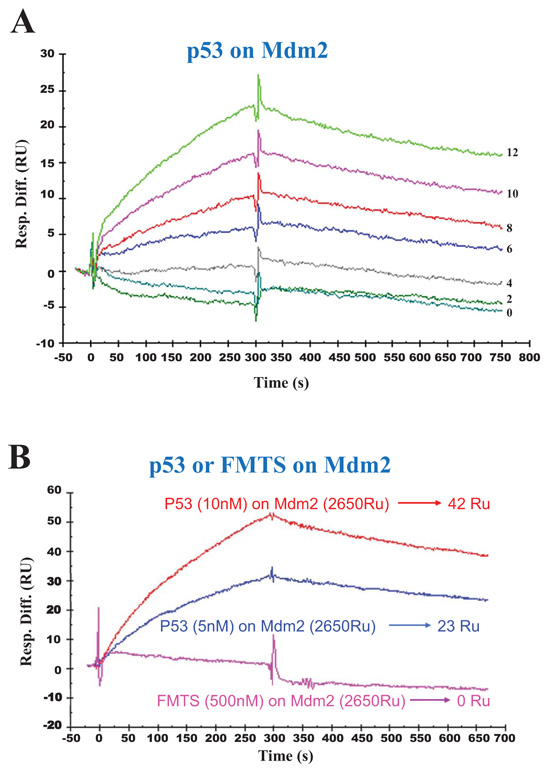
The His-tagged recombinant human Mdm2 was immobilized on the surface of CM5 sensor chip at low density resonance units (RU). (A): kinetics of interaction between Mdm2 and p53 at the concentrations of 0, 2, 4, 6, 8, 10, 12 nM. (B): demonstration of specific binding of p53 at the concentration of 5 nM or 10 nM on Mdm2 versus negative control (FMTS 500nM).
4. DISCUSSION
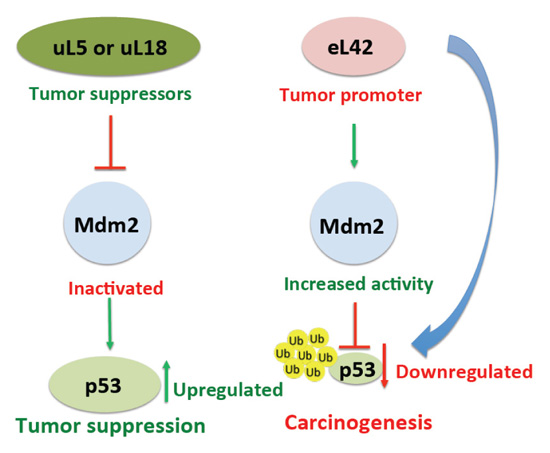
It was previously demonstrated that, following a nucleolar stress or a damage to DNA, certain ribosomal proteins are responsible for the regulation of the p53 level, either directly or indirectly through the rp-Mdm2-p53 axis [30-32]. In addition, the importance of the rp-Mdm2-p53 pathway in cancer development had been previously reported [30, 31]. To date, up to 16 rps have been identified to bind to Mdm2, particularly to the central acidic domain, and to regulate p53 activity [30]. It should be noted that almost all these rps exert a protective effect on p53 by inhibiting Mdm2 E3 ligase activity [30], each of them playing a different role in the regulation of the p53-Mdm2 axis [32]. For example, it was previously reported that rps uL5 and uL18 (formerly L11 and L5, respectively) negatively regulate Mdm2 by inhibiting p53 ubiquitination and degradation (Fig. 8 and [9, 10]). The case of rp eL42 is interesting in that this protein was found to be overexpressed in several cancer cells including human hepatocellular carcinoma, as well as in several human tumor cell-lines, and it was proposed that its overexpression might be related to tumor cell proliferation [14]. One explanation for the overexpression of eL42 in cancer cells is that it could negatively regulate the p53 protein, in contrast to the majority of the rps which were shown to exert a protective effect on p53. In the present report, we have addressed this question by studying the protein:protein interactions in the rp-Mdm2-p53 axis.
4.1. Interaction between rp eL42 and p53
In order to check the importance of eL42 in the rp-Mdm2-p53 axis, we have studied the interactions between eL42, p53 and Mdm2 by the co-immunoprecipitation technique on one hand, and by means of the Biacore assay, one the other hand. Regarding the co-immunoprecipitation data, it was shown that eL42 can bind to both p53 and Mdm2 (Figs. 2 and 3). In the Co-IP reaction of eL42 and p53, immunoprecipitation of eL42 allowed coimmunoprecipitation of p53 and vice versa (Figs. 2). These observations have been confirmed by kinetic measurements. The dissociation constant (KD) value of 1.22 x 10-8 M determined for the interaction between human eL42 and p53 (Fig. 4A and Table 1) would reflect a good binding affinity between these proteins. Moreover, the half maximal effective concentration (EC50) of p53 that directs half-maximal binding of the tumor suppressor onto the ribosomal protein was found equal to 2.2 x 10-9 M. On account of the 1:1 Langmuir interaction model, an EC50 in the nanomolar range would suggest that the interaction between eL42 and p53 is characterized by a strong binding affinity which argues for specific interactions. In principle, the values of KD and EC50 should be of the same order of magnitude in that they both reflect the strength of the eL42-p53 interaction. The difference between the value of KD (1.22 x 10-8 M) and that of EC50 (2.2 x 10-9 M) is likely to reflect a positive cooperativity binding mode between eL42 and p53, as suggested by the sigmoidal shape of the saturation curve in Fig. (4B). It is interesting to note that a positive cooperativity binding mode between eL42 and p53 would suggest that the interaction is of functional importance. Finally, a robust interaction in the nanomolar range is likely to trigger the sequestration of p53, especially in the case of overexpression of eL42, leading to an increase in proliferation by the inhibition of the tumor suppressor activity of p53. In this sense, eL42 would act as a cancer promoter, while its overexpression can be considered as a direct cause of cancer formation (Fig. 8).
4.2. Other Putative Roles for the Overexpression of eL42 in Cancer Cells
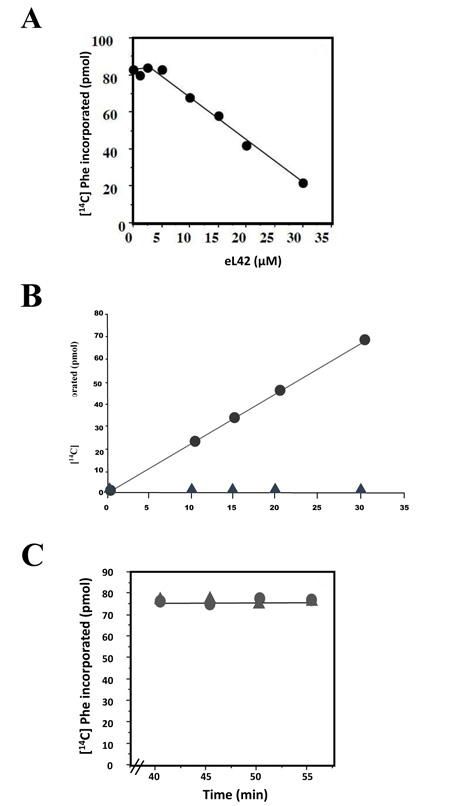
Another explanation for the overexpression of eL42 in cancer is that this rp essential for the elongation step of translation [13] could be overproduced to enhance the rate of the translation process in order to sustain the capacity of hyperproliferation of cancer cells. Thus, in addition to its extraribosomal role, the overexpression of eL42 would be favourable to cancer promotion in line with its ribosomal role in increasing the rate of translation elongation [13]. This proposition is consistent with the demonstration that Lys-55 residue of yeast eL42 was recently shown to be involved in the catalysis of peptide bond formation at the peptidyl transferase center of S. pombe ribosomes [13]. We addressed the question of an eventual ribosomal role of eL42 overexpression on the increase of the rate of translation elongation by measuring the poly(U)-dependent poly(Phe) synthesis activity of human 80S ribosomes in the presence of purified human recombinant eL42. As shown in Fig. (7A), not only the activity of the human 80S ribosomes is not increased in the presence of added eL42 protein, but it was decreased as a function of increasing concentrations of the protein. It was verified that, in the presence of a large amount (35 μM) of added eL42, the activity of the human 80S ribosomes was totally lost (Fig. 7B). We suspected that rp eL42 interferes with a component of the incubation mixture for the poly(U)-dependent poly(Phe) synthesis activity such as the tRNA substrate, because we had previously demonstrated that the interactions between eL42 and the tRNA molecules are characterized by strong binding affinities (in the nanomolar range) [22]. If this is the case, then the decrease in the poly(U)-dependent poly(Phe) synthesis activity would reflect the sequestration of the substrate [14C]Phe-tRNAPhe that would prevent the poly(U)-directed [14C]Phe incorporation into the poly(Phe) chain. The effect on activity of eL42 addition to the incubation mixture for poly(Phe) synthesis was also checked by following the activity of human 80S ribosomes when [14C]Phe incorporation had reached a plateau value prior to eL42 addition. Fig. (7C) shows that, despite the addition of a large amount (35 μM) of eL42, the activity of the human 80S ribosomes remained stable after having reached a plateau value. This result would support the suggestion that, when [14C]Phe incorporation was complete, the addition of eL42 could not provoke the decrease in the poly(U)-dependent poly(Phe) synthesis activity any more. Sequestration of tRNA by eL42 would have deleterious effects on cancer cells, because it would provoke the inhibition of the translation process by depriving the ribosomes of the tRNA substrates, thus slowing down cell growth.
At this stage, one can wonder if the only role of the overexpression of eL42 as a cancer promoter is the downregulation of p53. However, the following previous observations strongly support another role for the overexpression of eL42 in cancer cells: (i) in the crystallographic structure of S. cerevisiae 80S ribosome or of the 50S subunit of Haloarcula marismortui (Hma), cycloheximide, an antibiotic of the glutarimide family, and a strong inhibitor of translation in eukaryotic cells, had been shown to target the eL42 protein and to make contact with the lysyl residue (Lys-55 in S. cerevisiae and Lys-51 in Hma) [13]. This result argues for a critical role for this lysyl residue at the PTC of the eukaryal or the archaebacterial ribosomes; (ii) in addition, several yeast strains are known to exhibit resistance mutations among which is the P56Q mutation in eL42 that renders them resistant to cycloheximide [13]. The fact that cycloheximide inhibits protein biosynthesis by binding to Lys-55 of eL42, while the mutation P56Q renders some yeast strains resistant to cycloheximide, indicates that these two adjacent amino acid residues (Lys-55 and Pro-56) are involved in the binding and in the inhibitory effect of cycloheximide on the activity of the ribosome; (iii) moreover, in a previous study on S. pombe cells [33], have demonstrated that the protein Set13 is a specific methyltransferase responsible for monomethylation at lysine 55 in rp eL42. The Δset13 cells deprived of the methyltransferase displayed higher cycloheximide sensitivity than wild-type cells [33], suggesting that the binding of the antibiotic to the side chain of Lys-55 is more efficient in the absence of the methyl group than in its presence. This observation is in accordance with the fact that the eL42-K55R mutant cells (the Arg residue is non methylatable by the Set13 methyltransferase) were even more sensitive to cycloheximide than the Δset13 cells, and suggests at the same time that binding of cycloheximide to rp eL42 most probably proceeds by hydrogen bond formation between the amino group of the side chain of Lys or Arg residues and a carbonyl group of the glutarimide ring, as confirmed by the crystallographic data [15, 33]; (iv) we have previously demonstrated that the residue Lys-53 of rp eL42 is methylated to about 50% inside the human 80S ribosomes, similarly to the Lys-55 residue of eL42 from S. pombe 80S ribosomes [33, 34], while the non-methylated fraction of these critical residues was shown to contribute directly and actively to the 80S ribosome’s peptidyl transferase activity by promoting the course of the elongation cycle [13]. Taken together, these data demonstrate that methylation of Lys-53 of human rp eL42 and of Lys-55 of S. pombe eL42 plays a direct role in cycloheximide sensitivity, which is tightly linked to ribosomal function and to cell proliferation control [13, 33, 34]. In this regard, since the non-methylated fraction of the critical Lys-53 and Lys-55 residues was shown to contribute to the activity of the 80S ribosomes in human and in the yeast [33], respectively, it is reasonable to hypothesize that, following overexpression of rp eL42, the excess of the protein would be only slightly subjected to methylation, giving rise to the accumulation of a large amount of ribosomes containing non-methylated eL42 and, consequently, to an increase in activity and cell growth.
4.3. Interaction between rp eL42 and Mdm2
The interaction between eL42 and Mdm2 was studied by using the Mdm2 [1-350] fragment containing all the functionally important domains of the protein, such as the binding sites for proteins eL42 and p53, which makes it suitable for the study of protein:protein interactions in the cancer-pertinent rp.eL42-p53-Mdm2 pathway. Analysis of the kinetic of the interaction between eL42 and Mdm2 [1-350] indicated a KD value of 7.09 × 10-6 M (Fig. 4C and Table 1). As for the well-known and well-defined p53:Mdm2 complex, the kinetic of interaction indicated a KD value of 2.21 × 10-6 M (Fig. 4C and Table 1). It is interesting to note that the interaction between eL42 and Mdm2 or between p53 and Mdm2 were shown to be accounted for by KD values in the micromolar range, which are about 2-3 orders of magnitude higher than that (in the nanomolar range) of eL42 binding to p53. These results would suggest that the eL42:p53 complex is formed with stronger binding affinities than the p53:Mdm2 and eL42:Mdm2 complexes (see Table 1) which exhibit comparable binding affinities. There are several observations that can be made in this regard: (i) considering that Mdm2 is the enzyme (E) ubiquitin E3 ligase which ubiquitinates the p53 substrate (S) in order to direct it toward degradation by the proteasome [8, 9], the KD value of 2.21 × 10-6 M for the Mdm2:p53 (E:S or ES) complex is compatible with the affinity of formation of a classical Michaelis-Menten E:S complex. In fact, a KD (or KM) value in the micromolar range is the reflect of an interaction of medium intensity that would be favourable to the release of the product out of the active site of the enzyme through the activated ES# complex of the transition state, while a KD (or KM) value in the nanomolar range would reflect a strong interaction in the activated ES# complex at the transition state that would make it very slow the release of the ubiquitinated p53 protein from the catalytic site of Mdm2; (ii) the same observations can be applied to eL42, because this ribosomal protein had been previously shown to be ubiquitinated by Mdm2, similarly to p53 [35]; (iii) ubiquitination of both eL42 and p53 by Mdm2 is compatible with the fact that the two proteins have each a specific binding site on the ubiquitin E3 ligase Mdm2 (Fig. 1). In fact, the tumor suppressor p53 had been shown to bind to the N-terminal domain of Mdm2, while rp eL42 was supposed to bind to the central negatively charged acidic domain of Mdm2 [35]. The polyanionic character of this domain was assumed to be complementary to the highly positively charged eL42 (pI 10.59).
4.4. Effect of Pyridoxal 5'-phosphate (PLP) on the Interaction between eL42 and p53 Protein
Given the importance of the p53-Mdm2-MdmX loop in the initiation and development of tumors, several studies have identified small-molecules that can specifically target the individual protein molecules of this pathway to develop better anticancer treatments [36-45]. Pyridoxal 5'-phosphate (PLP) could represent one of such molecules, because it had been previously shown to exhibit anticancer properties [46, 47]. Interestingly, we have recently demonstrated that the ribosomal proteins eL42 and bL12 can be crosslinked with PLP in situ in human and E. coli ribosomes, respectively ([26] and unpublished data). The KD value of 2.75 x 10-4 M for the PLP-eL42 interaction determined in the present report is compatible with the limited surface of interaction existing between the small-molecule inhibitors and their target proteins. It is interesting to note that the KD value for the p53-eL42 interaction in the presence of PLP (1.08 x 10-7 M) is tenfold higher than the KD value obtained in its absence (1.22 x 10-8 M), suggesting that this small-molecule inhibitor is capable of perturbing the interaction between eL42 and p53. Moreover, in the presence of PLP, the sigmoidal shape of the saturation curve was by far less pronounced (Fig. 6D). Altogether, these results would suggest that, not only PLP is capable of inhibiting the interaction between eL42 and p53, but it also affects the positive cooperativity binding mode between these proteins.
| Proteins | Ligands | ka=kon [M-1S-1] |
kd=koff [S-1] |
KD= kd/ka [M] |
|---|---|---|---|---|
| eL42 | p53 | 7,18×103 | 8,78×10-5 | 1,22×10-8 |
| eL42 | PLP | 9,72 | 2,67×10-3 | 2,75×10-4 |
| eL42 | PLP-treated p53 | 2,36×103 | 2,55×10-4 | 1,08×10-7 |
| eL42 | Mdm2 [1-350] | 180 | 1,28×10-3 | 7,09×10-6 |
| Mdm2 [1-350] | p53 | 429 | 9,5×10-4 | 2,21×10-6 |
CONCLUSION
In this study, we have explored the potential of the large subunit ribosomal protein eL42 as a therapeutic target in the human cancer-pertinent rp.eL42-p53-Mdm2 pathway. By using the co-immunoprecipitation technique and the binding assays on Biacore, we have demonstrated that: (i) the ribosomal protein eL42, the tumor suppressor protein p53 and the ubiquitin E3 ligase Mdm2 interact with each other in a ternary rp.eL42:p53:Mdm2 complex. In particular, the interaction between eL42 and p53 is characterized by a strong binding affinity (KD value in the nanomolar range) that is likely to trigger the sequestration of p53 and the inhibition of its tumor suppressor activity, especially in the case of overexpression of eL42. Consequently, we propose that eL42 might be considered as a cancer promoter, while its overexpression might represent a direct cause of cancer formation through the downregulation of the tumor suppressor p53 (Fig. 8); (ii) the p53:Mdm2 and eL42:Mdm2 complexes exhibit comparable binding affinities in the micromolar range compatible with Mdm2 being the enzyme which ubiquitinates both the p53 and eL42 substrates. It is proposed that a KD (or KM) value in the micromolar range is the reflect of an interaction of medium intensity that would permit the release of the ubiquitinated p53 or eL42 proteins out of the catalytic site of Mdm2; (iii) pyridoxal 5'-phosphate (PLP), one of the active forms of vitamin B6 binds to eL42, and significantly inhibits the interaction between eL42 and p53, in accordance with the observation that vitamin B6 is associated with reduced risk of cancer. Altogether, our data suggest that perturbing the rp.eL42-p53-Mdm2 network might have implications for tumorigenesis and present opportunities for cancer therapy. In conclusion, the current understanding of the major mechanisms of p53 downregulation includes one that is likely to be triggered by its sequestration by eL42 when this ribosomal protein is overexpressed, and one that is well known and that is triggered by the proteasomal degradation of p53 mediated by its ubiquitination by Mdm2.
LIST OF ABBREVIATIONS
| rp | = Ribosomal Protein |
| eL42 | = Eukaryal or archaeal large subunit ribosomal protein L42 (formerly L42A or L42AB in yeast, L36a or L36a-like in human, or L44e in archaea) |
| FMTS | = Formyl-methionine-tRNA synthetase |
| PLP | = Pyridoxal 5'-phosphate |
| SPR | = Surface plasmon resonance |
| Co-IP | = Co-immunoprecipitation |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
We are grateful to the Ministry of higher education in Benin for supporting the PhD work of BA and HH (ref. 125/MESRS/CAB/DC/SGM/DRFM/DRH/REGIE). We gratefully thank the “Service de Coopération et d’Action Culturelle (SCAC)” of the French Ambassy in Cotonou (Bénin) for the financial support to BA and HH (ref. 0185 BEN B15 0022). We thank the West African Economic and Monetary Union (UEMOA, Ouagadougou, Burkina Faso) for the financial support to BA (ref. 06612/DDH/DESFP/0281).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We are indebted to Professor Thierry Soussi for the kind gift of the recombinant plasmids pGEXp53FL and pGEXMdm2, and for insightful advises.