All published articles of this journal are available on ScienceDirect.
Characterization of Carotenoid-protein Complexes and Gene Expression Analysis Associated with Carotenoid Sequestration in Pigmented Cassava (Manihot Esculenta Crantz) Storage Root
Abstract
Carotenoid-protein complex (CPC) was isolated from chromoplast-enriched suspensions of cassava storage root (CSR) using size exclusion chromatography and characterized. Peptide sequences (LC_MS/MS spectrum) obtained from CPC and their corresponding proteins were obtained using publically available databases. Small Heat Shock Proteins (sHSPs) were the most abundant proteins identified in the CPC. Western blot analysis showed that Fribrillin and Or-protein were present in chromoplast-enriched suspensions of yellow root but not in the complex or white root. Results from qRT-PCR helped identify an isoform of HSP21 possessing four single point mutations in the intense yellow CSR that may be responsible for increased sequestration of b-carotene.
INTRODUCTION
Plastid lipid-associated proteins (PAPs) and Carotenoid Associated Proteins (CAP) are general terms conferred to two classes of proteins accumulated at high levels in chromoplasts of plant cells [1,2] that are involved in the sequestration of hydrophobic compounds including lipids and carotenoids. These proteins are being investigated by several groups to establish the role of their parallel accumulation with carotenoids [3] and lipids of different types [4,5]. These investigations also include examining chromoplast formation [2,3,6], which specially focus on the Fibrillar type that forms a carotenoid-protein complex (CPC) within supramolecular structures where different carotenoid types are sequestered [2,7,8]. Most of this information was obtained from green tissues where carotenoid function is well documented in dissipating excess excitation energy by participating in non-photochemical quenching, which is essential in protecting the chloroplasts from photo-oxidative damage [9] and lipid changes that occur during leaf senescence due to interconvertion of chloroplasts into chromoplast [5]. Here we provide information on the characterization of carotenoid-protein complex isolated in cassava storage root that is a non green tissue.
Analysis of sequence data generated by LC-MS/MS is one of the classic PROTEOMICs technologies widely adopted for sequencing complex protein mixtures and predicting individual proteins [10]. Although erroneous protein identification is appreciated [11], the identification of individual proteins by LC-MS/MS is largely supported by statistical scoring systems 12, http://systemsbiology.org/re-search/software.html., which increase our confidence about protein sequence identification and homology matches to peptide sequences deduced from species specific EST databases, including the EMBRAPA cassava genome project database http://genoma.embrapa.br/genoma/, ESTIMA database http://titan.biotec.uiuc.edu/cgi-bin/ESTWebsite/-estima_start?seqSet=cassava, CIAT data base http://webapp.ciat.cgiar.org/biotechnology/bioinformatics.htm, Riken full-length cDNA database http://www.brc.riken.go.jp/lab/epd/Eng/catalog/pDNA.shtml and the Arabidopsis counterparts (ATG_ORTHOLOGUE) (http://www.arabidopsis.org/). These resources and other public databases facilitate identification of proteins and there corresponding gene sequences using validated peptide sequences derived from shotgun PROTEOMIC approaches. Here, we report results from combining chromoplast-enriched suspension preparation, protein-carotenoid complex separation in conventional size exclusion chromatography (SEC), protein separation by SDS-PAGE, and shotgun PROTEOMIC sequence techniques to identify proteins from carotenoid-protein complex present in cassava storage root.
MATERIALS AND METHODOLOGY
Plant Material and Tissue Preparation
Cassava Storage Root (CSR) from field-grown plants at EMBRAPA Genetic Resources and Biotechnology Research Unit (Brasilia, DF. Brazil) were used in this study. For protein extraction, quantification and protein blot analysis we used landrace BGM019 (intense yellow root) and the cv. IAC12.829 (white root). Protein sequence from carotenoid-protein complex was obtained from landrace BGM019. For gene expression analysis we used landrace BGM019 and cv. IAC12-829. Fresh storage roots (cylinders of 30-40 cm length x 4-6 cm diameters) were manually sliced and immediately frozen in liquid nitrogen, freeze-dried and stored at -80°C until used. For the chromoplast-enriched suspension studies, fresh intact storage roots were peeled off and processed immediately after harvest.
Protein Extraction, Quantification and Blot Analysis
One gram of freeze-dried CSR tissue was rehydrated and added with 2 ml of extraction buffer (pH 6.8, 5% SDS, 10% glycerol, 80 mM Tris, and 25 mM DTT), and 20 ml acetone and incubated at –20°C for more than 1h to obtain a protein precipitate free of pigments. After centrifugation (30000 rpm @ 4°C for 20 min) the supernatant was drained, the pellet dried by blowing N2 air and suspended with 5 ml extraction buffer (EB). The re-suspended solution was centrifuged again (30000 rpm @ 4°C for 10 min) to collect 1ml aliquot of the supernatant that was stored at –80°C until use. Aliquots (100 µl) were treated with DOC (0.15%) and TCA (12%) to precipitate soluble proteins. After centrifugation (13000 rpm @ 4°C for 20 min) the pellet was suspended in 100 µl of AB, and 50 µl was used for tissue protein estimation by the BCA Micro Kit method (PIERCE) according to the manufacturer protocol. Proteins from crude extract (CE), peak1 and 2 (30 µg each) were separated by 12% SDS-PAGE and blotted onto nitrocellulose as previously described [13]. Transblots were blocked with nonfat dry milk and incubated with primary antibody to Or-protein or Fibrillin-protein at a 1:200 dilution and secondary AP-conjugated goat-anti-rabbit IgG antibody (Bio-Rad) at 1:3000 dilution. Western Blue Stabilized Substrate (Promega) was used for chromogenic detection.
Preparation of Chromoplast Enriched Suspensions
Fresh CSR was harvested, washed with tap water, sliced, and homogenized with Homogenate Buffer (HB) adjusted to pH 8.2 (100 mM Tris-HCl, 8 mM EDTA, 10 mM KCl, 2 mM MgCl2, 400 mM sucrose, and 1 mM PMSF) using a household blender to obtain a paste that was filtered through three layers of cheesecloth. The filtrate was centrifuged (600 rpm @ 4oC for 30 min) and the resulting supernatant was centrifuged (2000 rpm @ 4oC for 30 min) a second time. The resulting pellet was suspended by brushing the surface with 50 mL HB, washed once with HB, followed by centrifugation (2000 rpm @ 4oC for 30 min). The final pellet was collected by surface brushing with 50 mL of suspension buffer (SB) adjusted to pH 6.8 (100 mM Tris-HCl, and 250 mM NaCl), named Non-denatured Chromoplast-Enriched Suspension (NDCES), and used for separation of carotenoid-protein complex and non-carotenoid associated proteins by Size Exclusion Chromatography (SEC).
Size Exclusion Chromatography of NDCES
Chromoplast-enriched suspensions were sonicated for 10 min and 5 mL was loaded onto a column (1 m length x 1.5 cm internal diameter) filled with a resin-bed of Sepharose CL 6B-200 and equilibrated with SB. Fractions (1 mL) were collected at a flow rate of 1.5 mL min-1 and OD readings @ 280 nm and 461 nm were recorded using a spectrophotometer (Spectra max Plus_384, Molecular Devices) using UV transparent microplate. Fractions from two observed peaks were pooled separately, named Peak 1 (fractions 33-49, carotenoid-protein complexc - CPC) and Peak 2 (fractions 108-122, non carotenoid-protein complex - NCPC), concentrated, and stored at –80°C until further analyzes.
HPLC Analysis of Peak 1 Carotenoids
Pooled fractions 33-49 from SEC were treated with acetone to isolate the carotenoid constituent, which was further separated on a C30 column according to [14] to identify the composition of carotenoids present in Peak 1.
Protein Preparation for LC_MS/MS Analysis
Pooled protein fractions from peaks 1 and 2 were separated by one-dimensional SDS_PAGE as previously described [10,15,16] and gels zones were excised and used for in-gel protein digestion with trypsin. Equivalent protein amount (100 mg) from both peak 1 and 2 were individually transferred to a siliconized eppendorf tube and dried in a speed-vac devise. The dried pellet was re-suspended in 200 mL of TCEP solution (10 mM) prepared fresh in ambicard and incubated at 56°C for one hour and cooled to room temperature. Proteins were then precipitated with 4 volume of acetone after incubation at -80°C for one hour. After centrifugation at maximum speed for 30 min at 4°C, the pellet was collected and added to 200 mL of Iodacetamide (55 mM) in ambicarb, and incubated for one hour at room temperature in the dark. The sample was incubated at -80°C for one hour with 4 volumes acetone and then centrifuged (maximum speed @ 4°C for 30 min). The resulting pellet was incubated with 100 mL of ambicarb by vortexing and sonicating for one hour. A trypsin solution (20 mg mL-1 in ambicarb) was added to have a protease:protein ration of 1:50, incubated at 37°C for the first two hours, after which an additional 100 mL of the same trypsin solution was added for incubation over night. After cooling to room temperature the sample was used for mass spectrometry analysis.
LC-MS/MS Analysis
After initial protein digestion, peptides were ionized, detected, isolated, fragmented and sequenced in a qTOF MALDI MS/MS. The operating software generated MS and MS/MS spectra, and the peptide sequences were determined using the PROTEOMIC facilities and software available at USDA/ARS-National Animal Disease Center in Ames, Iowa.
Bioinformatics Analysis of Peptide and Protein Sequence Data
Peptide sequences were established using a MASCOT server database (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF) and statistical validation as well as protein sequence matching identification. Peptide sequences that produced no protein hits were further analyzed using tblastx and translation tools available in HExPASy at Proteomics Server (http://www.expasy.org/) using four cassava EST databases located at CIAT (http://webapp.ciat.cgiar.org/biotechnology/bioinformatics.htm), ESTIMA (http://titan.biotec.uiuc.edu/cgi-bin/ESTWebsite/estima_start?seqSet=cassava), EMBRAPA (http://www.genoma.embrapa.br/), and RIKEN (http://www.brc.riken.go.jp/lab/epd/Eng/species/manihot.shtml). Finally, the protein fragment sequences derived from the EST matching results were mapped to an Arabidopsis protein database [http://www.arabidopsis.org] and to cassava PROTEOME and GENOME databases [http://www.phytozome.net/cassava]. Information on cassava proteome loci, for identified protein, and genome scaffold (for their corresponding gene) were obtained to characterize their location and distribution of identified proteins in the carotenoid-protein complex.
RNA Isolation, Quantification, and cDNA Synthesis
Total RNA was extracted twice using the phenol chloroform procedure described previously [17] and treated with RNAse free DNAse. Total RNA was quantified using the Quant-It RiboGreen RNA Kit according to the manufacturer (Molecular Probe). Total RNA (400 ng) was reverse transcribed in a 20 μL reaction volume using a SuperScript III Platinum Two-Step qRT-PCR Kit (Invitrogen). The reactions were terminated by heat inactivation and the cDNA products were treated with RNase H and stored at –20°C.
Gene-Specific Primers, Certified Standard and Housekeeping Gene Primers
Fluorogenic primers (FAM labeled LUX primer) and corresponding unlabeled primers were designed using the LUX Designer-Desktop version (Invitrogen) for each sequence of the cDNA fragment coding for genes as shown in Table S1 (see supplementary data). Standard certified primers for 18S ribosomal (Invitrogen Cat. 115HM-02) and Gus (Invitrogen Cat. 112H-02), and qPCR plasmid Standards (Invitrogen Cat. 11741-100) with Gus ORF were used as internal controls and quantitative standards to generate standard curves respectively. All primers were synthesized and purchased from Invitrogen.
Quantitative Real Time PCR Analysis
Gene sequences coding for proteins identified from the carotenoid-protein complexes were used to quantify transcript abundance in white and intense yellow CSR using (qRT-PCR). The qRT-PCR assays were performed in triplicate for each extraction using a Bio-Rad system (BioRad model iCycler). Ribosomal RNA (18S) was used as an internal control and PCR efficiency was evaluated using GUS certified primers to obtain a quantitative standard curve for absolute expression analysis of target transcript level in samples. Each experiment was replicated at least three times. Alternatively, a relative expression assay using MEC 1 (gene coding for Pt2L4 protein, 17) was used as an internal control. qRT_PCR products were sequenced at the EMBRAPA Genetic Resources and Biotechnology Genome facilities.
Data Analysis
To access the intrinsic property for the level of each transcript corresponding to the target gene in all samples, qRT-PCR amplification procedures consisted of absolute quantification expression using total RNA as normalizer. These methods provided an absolute quantitative standard curve for a dilution series based on the qPCR plasmid standards kit protocol (Invitrogene). Preliminary experiments with two unknown samples were performed to set up qRT-PCR amplification optimization conditions, including reliable exponential phase of amplification, qRT-PCR efficiency ranging from 90% to 105%, Ct threshold and baseline, and evaluation of each primer sets for target gene within an unknown sample. After setting up the optimized conditions, Ct values for all unknown samples and replications were interpolated from the standard curve estimating the level of transcript in unknown samples. In addition, a five point curve made with each amplicon was performed to obtain the efficiency of each primer (needed for the ΔΔct formula) and to know the range of amplification. After setting up the optimized conditions, Ct values for all unknown samples and replications were collected and normalized with housekeeping genes (internal controls) using the ΔΔct formula. Statistical parameters, such as mean and mean standard deviation, were performed in Excel.
RESULTS
Carotenoid-protein Complex Isolation and Characterization
Proteins from non-denatured chromoplast-enriched suspension (CES) present in white and intense yellow cassava, containing chromoplast-associated proteins (CAP) were separated by SEC Fig. (1A and 1B), respectively.
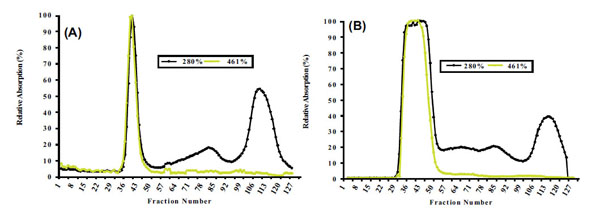
Size exclusion chromatography (SEC) profile for non-denature chromoplast enriched suspension from white (A) and intense yellow (B) cassava storage root. See resin specificity and performed conditions in MATERIALS AND METHODOLOGY.
Size exclusion chromatography profiles monitored with Abs461 (carotenoids) and Abs280 (proteins) indicated the presence of two major peaks. Peak 1 (fractions 33-49) eluted within the void volume of the column, as determined with blue dextran (MW 2000 kDa) and overlap between the carotenoid and protein profiles indicates that Peak 1 represents a carotenoid-protein complex (CPC) with an estimated size close to 400 KDa. Peak 2 (fractions 108-122) eluted later in the SEC profile and no observed overlap between carotenoids and proteins in Peak 2 suggests it represents non-carotenoid-associated proteins (NCPC). Proteins present in CPC and NCPC were separated by SDS-PAGE to visualize protein complexity and to fractionate proteins for LC_MS/MS analysis. From the gel image (see Supplementary data complementing Table S2 and S5), a greater number of proteins were observed in fractions forming NCPC than in fractions forming CPC. Peptide and protein sequences from Peaks 1 and 2 identified 83 and 106 peptides that assembled into 34 and 38 proteins respectively. Carotenoids from pooled fractions of CPC, separated by HPLC, indicate that β-carotene is the major carotenoid of the CPC isolated from both white (Fig. 2A) and intense yellow cassava (Fig. 2B) phenotypes. These results confirm the presence of carotenoid-protein complexes in cassava storage root that is similar to the one reported for carrot root [18-20] in terms of size, but different in the type of carotenoid and proteins types that includes β-carotene, α-carotene, and lutein as the major carotenoids and a single major protein of 18kDa in the CPC of carrot.
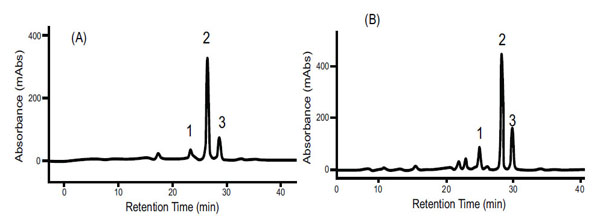
Carotenoid HPLC elution profile from the carotenoid-proteins complex (CPC) isolated by SEC as shown in Fig. (1). (A) spectrum from white cassava storage root. (B) Spectrum from intense yellow cassava storage root. (1) is Cis 9 β-carotene, (2) is All E β-carotene, (3) is Cis 13 β-carotene separated in a C30 column and identified by using external β-carotene standard, β-carotene absorption spectrum and retention time.
Peptide Sequence and Protein Identification in Chromoplast Enriched Suspension (CES)
Reverse-phase capillary chromatography (capillary LC) with quadrupole time-of-flight tandem spectrometry (nano-ESI Q-TOF MS/MS) was used with in gel tryptic digestion of proteins from CPC and NCPC. The LC-MS/MS peptide sequences were validated by MASCOT (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF) and used to identify proteins. The proteins sequences identified in CPC and NCPC are listed in the Supplementary data files in Table S2 and Table S5. A total of 65 proteins were identified, with 39 identified in NCPC and 26 in CPC. The majority of the proteins identified in both CPC (Fig.3A) and NCPC (Fig. 3B) are heat shock proteins (HSP) that are differentially distributed in the two fractions , which are the focus of discussion in this work. Out of 10 HSPs observed in both CPC and NCPC, four of them are present only in CPC (HSP15.7, HSP17.4I, HSP18.1I, and HSP26.5), four are present only in NCPC (HSP17.4, HSP17.6II, HSP26.5I and HSP70.1) and the remaining six are present in both fractions. This differential distribution of HSPs between these two fractions may indicate (1) differential properties of these HSPs to complex with carotenoids, (2) differential solubility in the SDS-PAGE gel separation procedure used in the present work, or (3) aggregation dynamics of a particular HSP in vitro or in vivo as reported by others [21].
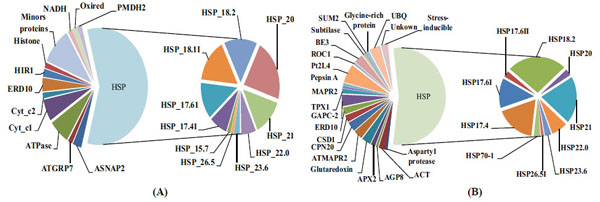
Distribution of proteins present in the carotenoid-protein complex (A) and non-carotenoid associated protein (B).
Occurrence of Fibrillin- and Or-protein in White and Intense Yellow Cassava
Protein-blot analysis was used to test for the presence of Fibrillin- and Or-proteins as markers for fibrillar structure of chromoplast occurrence in CSR as suggested in others plant systems [7,22], as well as chromoplast formation and differentiation [23,24] respectively. Fibrillin- (Fig. 4A) and Or-protein (Fig. 4B) are present in CES and NCPC of intense yellow cassava, but not in CPC isolated fractions or CES of the white cassava. These results suggest that the type of chromoplast structure accumulating carotenoid in cassava is of fibrillar-type . Since Fibrillin- and Or-proteins were not detected in the CES or in CPC of white cassava, these results also indicate that intense yellow root accumulate more chromoplasts than white roots of cassava. At the same time, these results suggest that further microscopic studies will be needed to confirm the form, number and the behavior of chromoplast in CSR, which was not addressed in the present work.
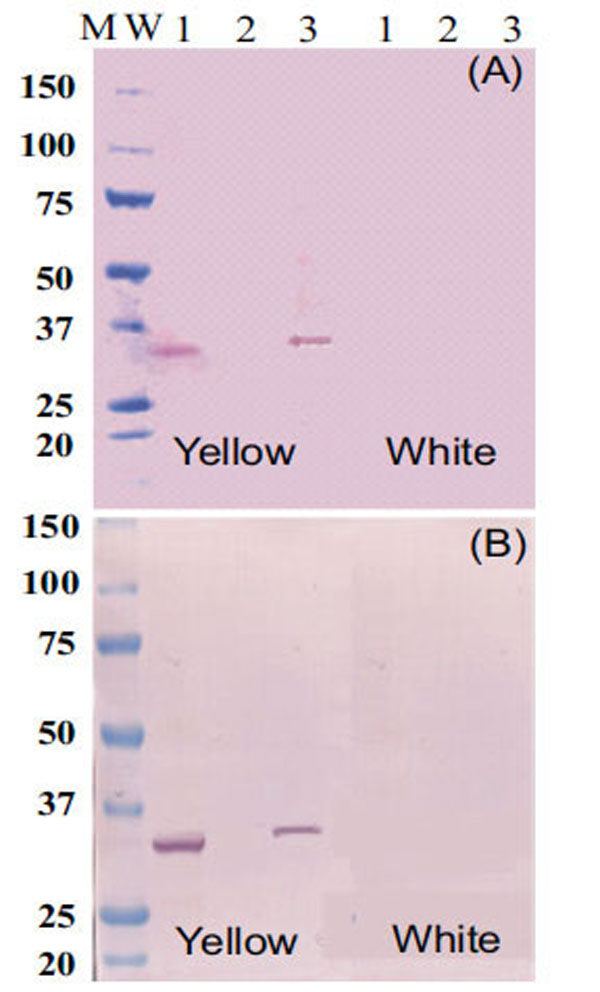
Western blot analysis of proteins (35µg/lane) extracts using antiserum raised against Fibrillin- (A) and Or-protein (B). Line 1 is chromoplast enriched suspension. Line 2 is total protein fraction from CPC. Line 3 is total protein fraction from NCAP identified in the profile of size exclusion chromatography as shown in Fig. (1).
Assign Assessed CAP Sequences to Cassava PROTEOME and GENOME
List of peptide and identified proteins sequences present in CPC are showed in Supplementary data in Table S2 and Supplementary data in Table S3 respectively. List of peptide and identified proteins sequences present in NCPC are showed in Supplementary data in Table S4 and Supplementary data in Table S5 respectively. The identified proteins were assigned to cassava PROTEOME and GENOME databases and results are summarized in Tables 1 to 4.
Identified Proteins in the Carotenoid-Protein Complex and their Distribution in the Cassava Genome
| Cassava Genome Scaffold | Score | E_Value | Protein Name Abbreviation |
|---|---|---|---|
| scaffold06948 | 971.5 | 0 | ACT7 |
| scaffold09150 | 836.2 | 0 | Actin |
| scaffold10689 | 291.6 | 4.9e-77 | Alpha_SNAP2 |
| scaffold04718 | 513.4 | 6.9e-144 | ATGRP7 |
| scaffold03604 | 1004.0 | 0 | ATPase |
| scaffold04355 | 921.0 | 0 | ATPase |
| scaffold03264 | 73.4 | 2.7e-11 | ATPsyn |
| scaffold04355 | 161.8 | 4.5e-38 | ATPsyn |
| scaffold04355 | 248.3 | 1.1e-63 | ATRZ-1A |
| scaffold06708 | 629.8 | 3.4e-179 | BE |
| scaffold08729 | 132.9 | 1.8e-29 | COX2 |
| scaffold06028 | 499.0 | 1.8e-139 | Cyto_c1 |
| scaffold06697 | 502.6 | 1.8e-140 | Cyto_c1 |
| scaffold05875 | 122.1 | 6.8e-26 | Dihy_acet |
| scaffold06708 | 747.9 | 0 | ERD10 |
| scaffold08053 | 302.4 | 2.2e-80 | HIR1 |
| scaffold12794 | 261.0 | 6.6e-68 | HIR1 |
| scaffold06656 | 244.7 | 4.4e-63 | Histone |
| scaffold05875 | 731.7 | 0 | HSP_17.4I |
| scaffold06512 | 1292.5 | 0 | HSP_17.6I |
| scaffold06908 | 949.9 | 0 | HSP_17.6I |
| scaffold06512 | 868.7 | 0 | HSP_18.1I |
| scaffold06908 | 1323.2 | 0 | HSP_18.1I |
| scaffold06512 | 868.7 | 0 | HSP_18.2 |
| scaffold06908 | 1323.2 | 0 | HSP_18.2 |
| scaffold08265 | 1545.0 | 0 | HSP_18.2 |
| scaffold06512 | 868.7 | 0 | HSP_20 |
| scaffold07543 | 1041.8 | 0 | HSP_21 |
| scaffold09151 | 1204.1 | 0 | HSP_22.0 |
| scaffold06701 | 1162.7 | 0 | HSP_23.6 |
| scaffold04151 | 881.3 | 0 | HSP_26.5 |
| scaffold04355 | 161.8 | 4.5e-38 | HSP17.6 |
| scaffold06512 | 358.4 | 4.0e-97 | HSP20 |
| scaffold06908 | 949.9 | 0 | HSP20 |
| scaffold08265 | 250.2 | 3.0e-65 | HSP20 |
| scaffold00069 | 978.7 | 0 | HSP21 |
| Cassava Genome Scaffold | Score | E_Value | Protein Name Abbreviation |
| scaffold06701 | 1162.7 | 0 | HSP23.6 |
| scaffold02264 | 149.2 | 2.3e-34 | MAB1 |
| scaffold00069 | 1038.2 | 0 | NADH |
| scaffold10097 | 468.4 | 3.2e-130 | OEP16 |
| scaffold07528 | 1402.5 | 0 | Oxireductase |
| scaffold07851 | 834.4 | 0 | PATL1 |
| scaffold06782 | 573.0 | 2.2e-161 | PMDH2 |
| scaffold06089 | 719.0 | 0 | RBP31 |
| scaffold00325 | 1031.0 | 0 | RRM_1 |
| scaffold07851 | 383.6 | 8.2e-105 | SEC14 |
| scaffold02892 | 798.4 | 0 | snRNP |
| scaffold12794 | 1123.0 | 0 | snRNP |
Identified Proteins in the Carotenoid-Protein Complex Assigned to Cassava Proteome Locus and their Distribution in Protein Families Assigned by Pfam Search
| Pfam | Manihot Locus | Protein Name Abbreviation | Score | E_Value |
|---|---|---|---|---|
| PF00022 | cassava4.1_033108m | ACT7 | 312.0 | 4.8e-85 |
| PF00022 | cassava4.1_033108m | Actin | 350.5 | 1.1e-96 |
| PF00515 | cassava4.1_013135m | ASNAP2 | 367.5 | 2.8e-102 |
| PF10559 | cassava4.1_006793m | ATPase | 194.5 | 3.5e-50 |
| PF00306 | cassava4.1_004726m | ATPsyn | 228.0 | 3.2e-60 |
| PF02806 | cassava4.1_001595m | BE | 1726.8 | 0 |
| PF02167 | cassava4.1_012354m | Cyt_c1 | 425.6 | 1.1e-119 |
| PF00198 | cassava4.1_004864m | Dihy_acet | 440.3 | 5.2e-124 |
| PF00257 | cassava4.1_015875m | ERD10 | 116.7 | 7.3e-27 |
| PF04716 | cassava4.1_017840m | RBP31 | 336.7 | 1.9e-92 |
| PF01145 | cassava4.1_013221m | HIR1 | 252.7 | 6.6e-68 |
| PF00125 | cassava4.1_019911m | Histone | 167.5 | 2.1e-42 |
| PF00011 | cassava4.1_019659m | HSP_15.7 | 225.7 | 1.6e-59 |
| PF00011 | cassava4.1_018038m | HSP_17.4I | 164.1 | 1.5e-61 |
| PF00011 | cassava4.1_018127m | HSP_17.6I | 261.2 | 3.8e-70 |
| PF00011 | cassava4.1_017871m | HSP_18.1I | 291.2 | 3.8e-79 |
| PF04969 | cassava4.1_017974m | HSP_18.2 | 266.9 | 9.5e-72 |
| PF00011 | cassava4.1_017871m | HSP_20 | 284.3 | 2.7e-77 |
| PF00011 | cassava4.1_015256m | HSP_21 | 396.4 | 1.1e-110 |
| PF04969 | cassava4.1_032436m | HSP_22.0 | 352.8 | 9.6e-98 |
| PF00011 | cassava4.1_016267m | HSP_23.6 | 382.9 | 1.3e-106 |
| PF00011 | cassava4.1_015518m | HSP_26.5 | 305.8 | 2.3e-83 |
| PF01423 | cassava4.1_019986m | snRNP | 58.9 | 2.8e-9 |
| PF02779 | cassava4.1_010415m | MAB1 | 237.3 | 1.6e-63 |
| PF02466 | cassava4.1_018513m | OEP16 | 256.1 | 9.4e-69 |
| PF00107 | cassava4.1_031433m | Oxireductase | 377.5 | 4.6e-105 |
| PF02866 | cassava4.1_010611m | PMDH2 | 612.1 | 0 |
| PF00076 | cassava4.1_012182m | RNP | 304.7 | 6.5e-83 |
| PF00076 | cassava4.1_019294m | RRM_1 | 161.4 | 4.5e-40 |
| PF03765 | cassava4.1_003845m | SEC14 | 309.3 | 6.2e-85 |
Identified Proteins in the Non-Carotenoid-Protein Complex and their Distribution in the Cassava Genome
| Cassava Scaffold | Total Score | E_Value | Protein Name Abbreviation |
|---|---|---|---|
| scaffold09150 | 839.9 | 0 | ACT7 |
| scaffold03602 | 206.9 | 2.2e-51 | ACT8 |
| scaffold03942 | 917.4 | 0 | AGP8 |
| scaffold06708 | 482.8 | 1.0e-134 | Amylase |
| scaffold00506 | 398.0 | 8.5e-109 | APX2 |
| scaffold06844 | 1067.1 | 0 | APX2 |
| scaffold00069 | 978.7 | 0 | Aspartyl protease |
| scaffold00847 | 906.6 | 0 | ATMAPR2 |
| scaffold06708 | 482.8 | 1.0e-134 | BE |
| scaffold00506 | 398.0 | 8.5e-109 | CPN20 |
| scaffold10114 | 462.9 | 2.3e-128 | CPN20 |
| scaffold04285 | 621.6 | 2.8e-176 | CSD1 |
| scaffold06705 | 1104.9 | 0 | CSD1 |
| scaffold06844 | 1067.1 | 0 | ERD10 |
| scaffold04953 | 627.1 | 1.3e-177 | GAPC |
| scaffold06948 | 720.8 | 0 | GAPC |
| scaffold07543 | 122.1 | 6.2e-26 | GAPC |
| scaffold03245 | 145.6 | 6.2e-33 | GAPC |
| scaffold07660 | 356.5 | 1.3e-96 | Glutaredoxin |
| scaffold07762 | 1103.1 | 0 | Glycine-rich protein |
| scaffold05255 | 919.2 | 0 | HSP17.4 |
| scaffold03651 | 1106.8 | 0 | HSP17.4 |
| scaffold01127 | 1005.8 | 0 | HSP17.4 |
| scaffold07859 | 1440.4 | 0 | HSP17.4 |
| scaffold06908 | 502.3 | 4.3e-141 | HSP17.4 |
| scaffold10469 | 1287.1 | 0 | HSP17.4 |
| scaffold12439 | 982.3 | 0 | HSP17.4 |
| scaffold09151 | 1204.1 | 0 | HSP17.4 |
| scaffold06512 | 1224.0 | 0 | HSP17.6I |
| scaffold05255 | 919.2 | 0 | HSP17.6I |
| scaffold10469 | 1287.1 | 0 | HSP17.6II |
| scaffold07859 | 1440.4 | 0 | HSP17.6II |
| scaffold06908 | 1285.3 | 0 | HSP18.2 |
| scaffold05875 | 731.7 | 0 | HSP18.2 |
| scaffold05255 | 919.2 | 0 | HSP18.2 |
| scaffold12439 | 1561.2 | 0 | HSP18.2 |
| scaffold08265 | 1545.0 | 0 | HSP18.2 |
| scaffold06908 | 1285.3 | 0 | HSP18.2 |
| scaffold06512 | 392.6 | 2.7e-107 | HSP20 |
| scaffold08265 | 250.2 | 3.0e-65 | HSP20 |
| scaffold00069 | 1038.2 | 0 | HSP21 |
| scaffold03241 | 872.3 | 0 | HSP21 |
| scaffold00467 | 596.4 | 1.1e-168 | HSP21 |
| scaffold02817 | 1178.9 | 0 | HSP21 |
| scaffold06908 | 502.3 | 4.3e-141 | HSP22.0 |
| scaffold09151 | 1204.1 | 0 | HSP22.0 |
| scaffold00069 | 978.7 | 0 | HSP23.6 |
| scaffold06512 | 868.7 | 0 | HSP23.6 |
| scaffold04151 | 881.3 | 0 | HSP26.5I |
| scaffold08265 | 571.2 | 3.8e-161 | HSP70-1 |
| scaffold08265 | 250.2 | 3.0e-65 | MAPR2 |
| scaffold04175 | 419.7 | 1.0e-115 | NAD binding |
| scaffold06598 | 378.2 | 3.4e-103 | Pepsin A |
| scaffold06844 | 1067.1 | 0 | Pt2L4 |
| scaffold07859 | 1440.4 | 0 | Pt2L4 |
| scaffold11638 | 1393.5 | 0 | ROC1 |
| scaffold08559 | 903.0 | 0 | Stress-inducible |
| scaffold00467 | 520.7 | 6.1e-146 | Subtilase |
| scaffold11661 | 414.3 | 3.4e-114 | SUM2 |
| scaffold00847 | 780.3 | 0 | TPX1 |
| scaffold10669 | 699.2 | 0 | UBQ10 |
| scaffold12498 | 558.5 | 2.3e-157 | UBQ2 |
| scaffold03055 | 1043.6 | 0 | UBQ2 |
| scaffold10669 | 699.2 | 0 | UBQ3 |
| scaffold00069 | 978.7 | 0 | UBQ6 |
| scaffold07591 | 749.7 | 0 | Unkown |
Identified Proteins in Non-carotenoid-protein Complex Assigned to Cassava Proteome Locus and their Distribution in Protein Families Assigned by Pfam Search
| Pfam | Cassava Locus | Protein Name Abbreviation | Total Score | E_Value |
|---|---|---|---|---|
| PF00022 | cassava4.1_033108m | ACT7 | 371.3 | 3.4e-103 |
| PF00022 | cassava4.1_033108m | ACT8 | 448.7 | 1.5e-126 |
| PF02806 | cassava4.1_001595m | AMYLASE | 255.8 | 6.2e-69 |
| PF00141 | cassava4.1_014643m | APX2 | 484.6 | 3.8e-137 |
| PF03489 | cassava4.1_005735m | Aspartyl protease | 517.7 | 4.6e-147 |
| PF00173 | cassava4.1_019974m | ATMAPR2 | 203.8 | 6.6e-53 |
| PF00166 | cassava4.1_014439m | CPN20 | 461.5 | 3.2e-130 |
| PF00080 | cassava4.1_018289m | CSD1 | 278.9 | 1.8e-75 |
| PF00257 | cassava4.1_015875m | ERD10 | 188.7 | 3.7e-48 |
| PF02469 | cassava4.1_031631m | Fasciclin | 323.2 | 4.5e-89 |
| PF00044 | cassava4.1_011175m | GAPC | 402.5 | 1.6e-112 |
| PF00044 | cassava4.1_011176m | GAPC | 287.0 | 6.3e-78 |
| PF00044 | cassava4.1_011326m | GAPC | 394.4 | 2.9e-110 |
| PF00044 | cassava4.1_011366m | GAPC | 634.4 | 0 |
| PF00462 | cassava4.1_019777m | glutaredoxin | 203.0 | 7.9e-53 |
| PF01277 | cassava4.1_016619m | glycine-rich protein | 243.4 | 7.0e-65 |
| PF00011 | cassava4.1_015256m | HSP17.4 | 403.3 | 9.5e-113 |
| PF00011 | cassava4.1_016281m | HSP17.4 | 302.8 | 1.5e-82 |
| PF04969 | cassava4.1_017267m | HSP17.4 | 295.0 | 9.6e-80 |
| PF00011 | cassava4.1_017871m | HSP17.4 | 291.2 | 3.8e-79 |
| PF00011 | cassava4.1_018066m | HSP17.4 | 241.1 | 2.4e-64 |
| PF00011 | cassava4.1_018093m | HSP17.4 | 281.6 | 3.8e-76 |
| PF00011 | cassava4.1_018121m | HSP17.4 | 305.4 | 1.4e-83 |
| PF00011 | cassava4.1_018127m | HSP17.4 | 261.2 | 4.0e-70 |
| PF00011 | cassava4.1_018158m | HSP17.4 | 267.7 | 5.5e-72 |
| PF00011 | cassava4.1_018376m | HSP17.4 | 295.8 | 1.0e-80 |
| PF00011 | cassava4.1_017871m | HSP17.6I | 291.2 | 3.8e-79 |
| PF00011 | cassava4.1_018127m | HSP17.6I | 261.2 | 3.8e-70 |
| PF00011 | cassava4.1_018131m | HSP17.6I | 167.2 | 2.1e-56 |
| PF00011 | cassava4.1_018158m | HSP17.6II | 267.7 | 5.5e-72 |
| PF00011 | cassava4.1_017871m | HSP18.2 | 291.2 | 3.8e-79 |
| PF00011 | cassava4.1_018038m | HSP18.2 | 164.1 | 1.5e-61 |
| PF00011 | cassava4.1_018066m | HSP18.2 | 241.1 | 2.4e-64 |
| PF00011 | cassava4.1_017871m | HSP20 | 291.2 | 3.8e-79 |
| PF00011 | cassava4.1_018131m | HSP20 | 235.0 | 3.7e-62 |
| PF00011 | cassava4.1_015256m | HSP21 | 396.4 | 1.1e-110 |
| PF00011 | cassava4.1_018066m | HSP21 | 241.1 | 2.4e-64 |
| PF00011 | cassava4.1_017871m | HSP22.0 | 291.2 | 3.8e-79 |
| PF00011 | cassava4.1_018066m | HSP22.0 | 241.1 | 2.4e-64 |
| PF00011 | cassava4.1_015256m | HSP23.6 | 396.4 | 1.1e-110 |
| PF00011 | cassava4.1_015518m | HSP23.6 | 305.8 | 2.3e-83 |
| PF04969 | cassava4.1_017267m | HSP23.6 | 295.0 | 9.6e-80 |
| PF04969 | cassava4.1_017974m | HSP23.6 | 266.9 | 9.5e-72 |
| PF04969 | cassava4.1_022774m | HSP23.6 | 226.9 | 1.1e-59 |
| PF04969 | cassava4.1_032436m | HSP23.6 | 352.8 | 9.6e-98 |
| PF00012 | cassava4.1_003331m | HSP70-1 | 194.5 | 9.2e-59 |
| PF00012 | cassava4.1_003343m | HSP70-1 | 379.0 | 3.0e-105 |
| PF05193 | cassava4.1_005898m | pepsin A | 274.2 | 1.3e-73 |
| UNKOWN | cassava4.1_017439m | Pt2L4 | 57.0 | 1.8e-8 |
| PF00160 | cassava4.1_017662m | ROC1 | 323.9 | 6.3e-89 |
| PF00515 | cassava4.1_004441m | stress-inducible | 305.8 | 1.3e-83 |
| PF05922 | cassava4.1_019197m | subtilase | 221.5 | 2.9e-58 |
| PF00240 | cassava4.1_019468m | SUM2 | 117.1 | 2.1e-27 |
| PF00578 | cassava4.1_017973m | TPX1 | 322.0 | 1.9e-88 |
| PF00240 | cassava4.1_019139m | UBQ10 | 247.3 | 4.5e-66 |
| PF07708 | cassava4.1_009650m | UBQ2 | 313.9 | 2.6e-160 |
| PF11976 | cassava4.1_018181m | UBQ2 | 266.5 | 6.5e-72 |
| PF00240 | cassava4.1_019139m | UBQ3 | 247.3 | 4.6e-66 |
| UNKOWN | cassava4.1_015690m | Unkown | 287.0 | 1.1e-77 |
It was observed that 30 identified proteins in CPC were distributed in 28 cassava-protein loci and 19 families while 33 proteins in NCPC are distributed in 44 loci and 25 families. The HSP group individuals for CPC are located in 9 loci. and distributed in 2 protein families. The gene coding for those proteins are distributed in 30 scaffolds for the CPC and 43 for the NCPC in the cassava GENOME. For the case of HSP, the genes coding for 10 HSP identified in CPC and in NCPC showed different distribution pattern, being 11 scaf folds for the CPC group and 15 scaffolds for the NCPC group. These results contribute to the overall observation about the functional complexity of HSPs in plants as well as the distribution and arrangement of their genes in the cassava genome.
Gene Expression Analysis
Expression of two genes, one related to chromoplast structure definition (Fibrillin gene) and another related to chromoplast development (Or-gene coding for Or-protein), were analyzed to gain insights on abundance and formation of chromoplast in CSR. Four identified gene sequences coding for proteins in the CPC (HSP18.1, HSP17.6, HSP17.4 and HSP21) were used to search for their possible roles in differential sequestration of β-carotene in white and intense yellow genotypes. Transcript levels for the above listed genes were more abundant in the intense yellow than in white CSR (Fig. 5) and followed the pattern of carotenoid accumulation.
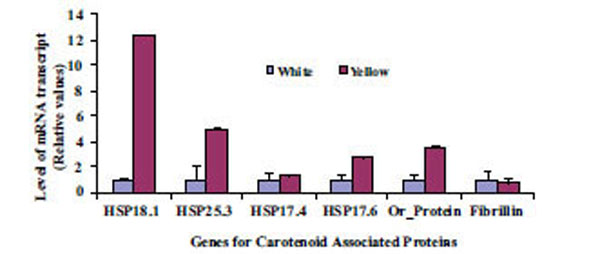
Differential levels of transcript for genes coding for a set of proteins related to carotenoid accumulation in white (A) and intense yellow (B). Relative values expressed in relation to white roots.
The dissociation temperature curve patterns shown in Fig. (6) of the qRT-PCR amplification of the four genes listed above indicates a change in nucleotide coding sequence for HSP21 (Fig. 6D), which is associated with high amounts of carotenoid accumulation in the intense yellow landrace. The overall observation about the functional complexity of HSPs in plants as well as their distribution and arrangements of their genes in the cassava genome. Indeed, the sequence of the corresponding amplified fragment (Fig. 7) revealed four single point mutations in the gene coding for this protein that may favor carotenoid sequestration in the CSR of intense yellow phenotype.
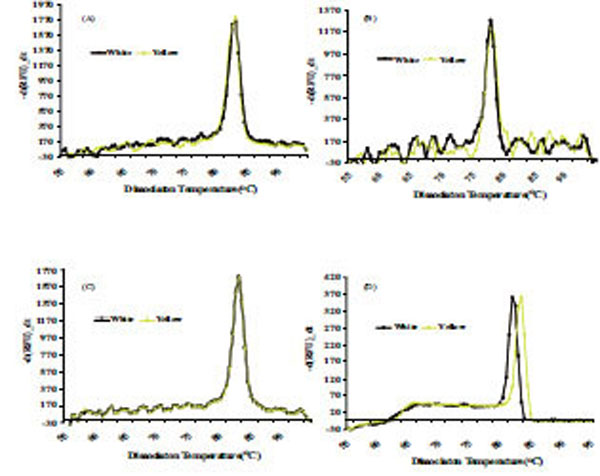
Dissociation curve derived from qRT-PCR amplification for genes coding for Fibrillin (A), Or-protein (B), CAP1 (C), CAP2 (D), present in CPC of white and intense yellow cassava storage root.
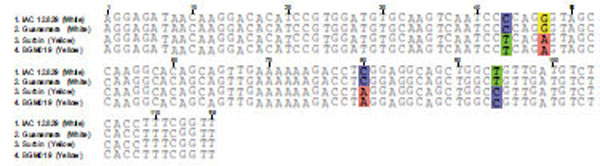
Sequence comparison for the amplicon resulting from qRT-PCR of HSP21 from white and intense yellow cassava storage root.
In addition, these results corroborate those observed by protein blot analysis where Fibrillin- and Or-protein support a role for enhancement of protein carotenoid sink capacity (i.e. chromoplast formation and number) as well as a possible specific role for HSP21 in the carotenoid sequestration mechanism.
DISCUSSION
Carotenoid-protein complex (CPC) formation in specialized supramolecular structures inside the chromoplast has been associated with several molecular functional interactions [1,24], including sequestration of specific carotenoids that aid in driving the carotenoid synthesis pathway towards completion. The characterization of CPC protein composition in chromoplast of cassava storage root (CSR) identified HSPs as the major type of proteins present in the complex. Further, specific markers for proteins (Fibrillin) used in this study indicate chromoplasts of intense yellow cassava storage root are of fibrillar type capable of chromoplast differentiation [25-27] and carotenoid accumulation [25]. Results from this study also suggest that a specific haplotype of HSP21 could explain the differential carotenoid accumulation in intense yellow cassava landraces.
HSPs are classes of proteins that are unique, abundant and diverse in plants, especially in non-green tissue [28,29]. Higher plants have at least 20 small Heat Shock Proteins (sHSPs) and some species may have up to 40 different sHSPs [30]. The sHSPs are involved in response to environmental stress such as heat, cold, drought and salinity stress [31,32]. In the absence of environmental stress, sHSPs expression in plants are restricted to certain stages of development such as embryogenesis, seed germination, pollen development, and fruit maturation [33,34]. In the case of CSR, we observed that the proteins forming the CPC in root chromoplast are mainly composed of the sHSP family with variable levels of expression, depending on the type of carotenoid accumulating genotype. For instance, transcripts for HSP18.1 and HSP21 are more abundant in intense yellow cassava storage root where values of β-carotene accumulation were 100x more than in white root. While HSP18.1 may be responsible for the enhancement of total carotenoid accumulation, HSP21 may also play a role in the specificity to β-carotene accumulation throughout the sequestration mechanism as observed in tomato [35]. In addition, a new heteroduplex double strand cDNA sequence for the gene coding for HSP21 was observed, suggesting that an isoform of this protein is coded by a different gene haplotype possesing four single point mutations in the sequence of the amplicon (Fig. 7) that may be responsible for the sequestration of high β-carotene in intense yellow CSR. In other plant systems, the presence of a specific HSP21 protein, has been speculated to act as an antioxidant under oxidative stress [36] or as an enhancer of carotenoid accumulation in tomato fruit of a transgenic plant [35]. More in-depth research on the genotypes enriched with special carotenoids from other cassava landraces, will have a larger impact on carotenoid genetic improvement in new varieties and strengthen our knowledge of protein content enhancement and carotenoid regulation in non-green tissue.
Fibrillin has been found in a number of plants and plays a structural role in carotenoid-storing fibrillar type chromoplasts [37]. This prompted us to examine the presence of fibrillin in CSR, which could help define the type of ultramolecular structure of chromoplast present in CSR. Here we demonstrated that fibrillin antibody recognized a putative cassava fibrillin protein in CES of intense yellow but not in white cassava storage root. This evidence supports the presence of carotenoid-fibril type in CSR, rather than other type of structures observed in roots such as carrot [18] that also store high amounts of carotenoids. Based on the discovery of Or-protein and its cellular function in inducing formation of chromoplasts [25,38], we used Or-protein antibody to recognize this protein in CSR as a possible indicator of the differential number of chromoplasts between intense yellow and white CSR. Indeed, Or-protein was observed in CES of intense yellow but not in white root. This evidence indicates the potential for more chromoplasts present in intense yellow CSR creating a metabolic sink for carotenoid accumulation, similar to that observed in transgenic potato tuber over-expressing the Or-gene that accumulate greater levels carotenoids [39].
CONCLUSION
In conclusion, this study provides evidence that the carotenoid-protein complex, in cassava storage root, is enriched with HSPs that interact with different carotenoids, depending on the genetic background of cassava landrace. The presence of Fibrillin- and Or-protein may confirm the fibrillar type of chromoplast and intense chromoplast formation in storage root of intense yellow compared to white cassava CSR. The novel allele of HSP21, identified in the present work, may be an important tool for genetic manipulation of protein content in cassava storage root contributing to a natural sink for protein and carotenoid accumulation in intense yellow CSR. Further elucidation of the roles of HSP21 in binding specifically with β-carotene or in inducing accumulation of carotenoid in CSR would provide a significant contribution to our understanding of the molecular machinery controlling carotenoid accumulation in non-green tissues.
CONFLICTS OF INTEREST
There are no conflicts of interest and the authors agree not to withdraw their manuscript at any stage prior to publication.
AKNOWLEDGEMENTS
Financial support from EMBRAPA/LABEX-US, in collaboration with the USDA-ARS, Red River Valley Agricultural Research Center in Fargo, ND, is acknowledged. Special acknowledges are also extended for the financial support provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (grant number 680.410/01-1), the National Biotechnology Program of EMBRAPA (grant number 06.03.02.058), International Atomic Energy Agency (grant number BRA_13188), State Administration of Foreign Experts Affairs, China (grant number Y20100-326001) as well as the KLUGRTC, MA/CATAS_TCGRI from China (grant number KFKT_2010_01.
SYMBOLS AND ABBREVIATIONS
URL ADDRESSES OF SUPPORT ANALYTICAL SOFTWARE AND DATA BASE
| MASCOT server | =http://www.matrixscienc.com/cgi/search_form.pl?FORMER=2SEARCH=PMF |
| ExPASy | =http://www.expasy.org/ |
| NCBI | =http://www.ncbi.nlm.nih.gov/sites/entrez?db=nucestcmd=searchterm=carvalho%20 cassava |
| CIAT data base | =http://webapp.ciat.cgiar.org/biotechnology/bioinformatics.htm |
| ESTIMA data base | =http://titan.biotec.uiuc.edu/cgi-bin/ESTWebsite/estima_start?seqSet=cassava |
| EMBRAPA data base | =http://www.genoma.embrapa.br/ |
| RIKEN data base | =http://www.brc.riken.go.jp/lab/epd/Eng/species/maniho.shtml |
| Cassava genome data base | =http://www.phytozome.net/cassava |
| Arabidopsis data base | =http://www.arabidopsis.org/ |
| Pfam Server | =http://pfam.sanger.ac.uk/ |
REFERENCES
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]


