All published articles of this journal are available on ScienceDirect.
Armadillo Repeat Containing 8α Binds to HRS and Promotes HRS Interaction with Ubiquitinated Proteins
Abstract
Recently, we reported that a complex with an essential role in the degradation of Fructose-1,6-bisphosphatase in yeast is well conserved in mammalian cells; we named this mammalian complex C-terminal to the Lissencephaly type-1-like homology (CTLH) complex. Although the function of the CTLH complex remains unclear, here we used yeast two-hybrid screening to isolate Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) as a protein binding to a key component of CTLH complex, Armadillo repeat containing 8 (ARMc8) α. The association was confirmed by a yeast two-hybrid assay and a co-immunoprecipitation assay. The proline-rich domain of HRS was essential for the association. As demonstrated through immunofluorescence microscopy, ARMc8α co-localized with HRS. ARMc8α promoted the interaction of HRS with various ubiquitinated proteins through the ubiquitin-interacting motif. These findings suggest that HRS mediates protein endosomal trafficking partly through its interaction with ARMc8α.
INTRODUCTION
Fructose-1,6-bisphosphatase (FBPase) is a key enzyme in gluconeogenesis in Saccharomyces cerevisiae. FBPase is synthesized when cells are grown in poor glucose media. When glucose-starved yeast cells are given fresh glucose media, FBPase is rapidly degraded by vacuole-dependent and proteasome-dependent pathways [1]. FBPase is targeted from the cytosol to intermediate vesicles and then to the yeast vacuole for the degradation in vacuole-dependent manner [2]. Yeast genes, the products of which are involved in the vacuole-dependent FBPase degradation, have been genetically isolated and named vacuolarimport and degradation-deficient (vid) genes [3-6]. On the other hand FBPase degradation on glucose addition requires multiubiquitin conjugate and is dependent on 26S proteasome [7]. Regelmann J. and his co-workers identified nine yeast glucose-induced degradation (gid) genes, the products of which are required for proteasome-dependent degradation of FBPase [8]. Thus the genes involved in these two alternative mechanisms have been reported in detail. Interestingly, gid1, gid4, and gid5 are identical to vid30,vid24, and vid28, respectively [8]. Recently, systematic mass spectrometric searches revealed that Gid1p/Vid30p, Gid2p, Gid4p/Vid24p, Gid5p/Vid28p, Gid7p, Gid8p, and Gid9p form a large protein complex [8, 9]. Therefore, the complex in yeast plays important roles in both vacuole- and proteasome-dependent FBPase degradation.
Recently, our group purified a novel complex containing Ran-binding protein in microtubule organizing center (RanBPM) [10], which is thought to be a scaffolding protein in the human immune and nervous systems [11]. We revealed that the complex comprised RanBPM, muskelin, p48 Erythroblast macrophage attacher-like protein (EMLP), p44 C-terminal to the Lissencephaly type-1-like homology (CTLH), Two-hybrid associated protein (Twa1), and the novel armadillo proteins, Armadillo repeat containing 8 (ARMc8) α and β from extracts of Human embryonic kidney (HEK) 293 cells [10]. Most of the components, except ARMc8α and ARMc8β, possess a Lissencephaly type-1-like homology (LisH) / CTLH motif, which is present in proteins involved in microtubule dynamics, cell migration, nucleokinesis, and chromosome segregation [12]. We named the complex in mammalian cell CTLH complex. Interestingly, our homology search of each amino acid sequence revealed that RanBPM, muskelin, p48EMLP, p44CTLH, and ARMc8α are human homologs of Gid1p/Vid30p, Gid7p, Gid9p, Gid2p, and Gid5p/Vid28, respectively. This finding led us to hypothesize that the human CTLH complex plays important roles in lysosome- and proteasome-dependent proteolysis.
ARMc8 proteins are novel armadillo-repeat containing proteins. The armadillo-repeat is an approximately 40 amino acid long tandemly repeated sequence motif which was originally identified in the Drosophila segment polarity gene product [13]. Armadillo-repeat containing proteins are well conserved through eukaryotes and are found in various proteins involved in diverse cellular functions including intracellular signaling and cell-cell contact regulation by interacting with a number of specific binding partners [14]. β-Cateninn [15], importin-α [16] and Adenomatous Polyposis Coli [17] are also armadillo-repeat containing proteins and regulate a variety of cellular process. To clarify the function of the CTLH complex, we focused on the novel armadillo proteins ARMc8α and β. We previously reported that the CTLH motif is a binding domain for ARMc8α and β [10]. Therefore, we hypothesized that ARMc8α and β would be key molecules for the functioning of the CTLH complex.
Here, we used a yeast two-hybrid screening assay and a human skeletal muscle cDNA library to search for proteins that might interact with ARMc8α; the C-terminal sequence of ARMc8α was used as bait. The endosomal protein Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) was isolated as a binding partner for ARMc8α. HRS is known to play an important role in the endosomal trafficking of membrane proteins such as epidermal growth factor receptor (EGFR) [18], Drosophila signaling receptors [19], vascular endothelial growth factor receptor (VEGFR) 2 [20], and Toll-like receptor 4 [21]. Monoubiquitination provides a signal for the sorting of these membrane proteins into the multi-vesicular body (MVB), which is targeted to lysosome [22]. In this process, HRS plays an essential role in recognizing the monoubiquitinated proteins via ubiquitin interacting motif (UIM) and regulates the endosome/ lysosome-dependent degradation of the membrane proteins [22].
Here, we characterize the association between ARMc8α and HRS. We also reveal that ARMc8α partly regulates the lysosomal trafficking of ubiquitinated proteins through its interaction with HRS.
MATERIALS AND METHODS
Antibodies
An anti-EEA1 antibody, anti-Omni probe, and anti-Ub antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). FLAG monoclonal antibody (M2) and an anti-α-tubulin monoclonal antibody were obtained from Sigma (St. Louis, MO), and T7·Tag monoclonal antibody from Novagen (Madison, WI). Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit IgG antibodies were obtained from GE Healthcare (Piscataway, NJ). Alexa Fluor 488 anti-mouse and anti-rabbit IgG antibodies and Alexa Fluor 594 anti-mouse and anti-rabbit IgG antibodies were purchased from Molecular Probes (Eugene, OR). Polyclonal rabbit antibody against human ARMc8 was made as previously described [10]. An anti-HRS antibody was purchased from Alexis Biochemicals (San Diego, CA).
Yeast Two-Hybrid Screening
The Matchmaker Gal4 Two-Hybrid System 3 (Clontech Laboratories, Mountain View, CA) was used on a human muscle cDNA library (Clontech Laboratories). For the screening we employed a widely used interaction mating protocol for yeast two-hybrid systems. AH109 yeast cells were transformed with the bait construct pGBKT7-ARMc8α (268–648), and the transformants were mixed for mating with yeast strain Y187 cells pre-transformed with the cDNA library fused to the pACT2 vector. Then the diploid transformants were selected on SD/-Ade/-His/-Leu/-Trp/X-α-Gal plates. The plasmids in positive clones were isolated and subjected to DNA sequencing analysis. Each piece of sequence data was compared with a database (BLAST server of the National Center for Biotechnology Information, Bethesda, MD).
Western Blotting and Immunoprecipitation Assay
The cells were washed with phosphate-buffered saline (PBS) twice, collected, then lysed in RIPA buffer [50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 1.0% Nonidet P-40] supplemented with protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). After incubation on ice for 20 min the lysate was centrifuged at 20 000 ×g for 30 min and the supernatants were collected. These samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the separated proteins were transferred onto Immobilon-P membranes (Millipore, Temecula, CA). After blocking, the membrane was probed with a primary antibody and with an adequate secondary HRP-conjugated antibody, and then visualized by using ECL Western Blotting Reagents (GE Healthcare) and an LAS-3000 mini luminescent image analyzer (FujiFilm, Tokyo, Japan). For immunoprecipitation assay, the cells were lysed in RIPA buffer in the same way described above. The lysate was then centrifuged at 20 000 ×g for 30 min and the supernatant was incubated and stirred with protein G-Sepharose (GE Healthcare) preliminarily conjugated with an appropriate antibody for 18 h at 4 °C. The immunoprecipitates were washed 4 times with RIPA buffer, and the bound proteins were eluted in RIPA buffer containing 2× Laemmli sample buffer by boiling the beads for 5 min, then analyzed by SDS-PAGE followed by immunoblotting.
Immunofluorescence Microscopy
HEK293 cells were cultured on glass coverslips for 24 h and transfected with an appropriate plasmid DNA. Immunofluorescence microscopy was performed as described previously [23].
Construction of Expression Vectors
To construct pCR-HRS, the full-length open reading frame (ORF) of HRS was amplified by a standard PCR technique using KOD polymerase (Toyobo, Osaka, Japan), the oligonucleotide primers (sense: 5'-GGATCCATGGGGC GAGGCAGCGGCAC; antisense: 5'-GGATCCTCAGTCGA ATGAAATGAGCT), and a human liver cDNA library (Clontech laboratories) as a template. The PCR product was ligated into the pCR-Blunt II TOPO (Invitrogen, Carlsbad, CA). To construct pT7-HRS, pCR-HRS was digested with BamH1 and the ORF was subcloned into pT7-Mock. To construct pACT2-HRS, the ORF of HRS was amplified by using the oligonucleotide primers (sense: 5'-GGATCCGAA TGGGGCGAGGCAGCGGCAC; antisense: 5'-GGATCCTC AGTCGAATGAAATGAGCT) and pT7-HRS as a template. The resulting product was ligated into the pCR-Blunt II TOPO; the pCR-HRS was digested with BamH1 and the ORF was subcloned into pACT2-Mock. To obtain HRS deletion constructs (p144–777, p241–777, p335–777, p391–777, p1–245, p1–549), each deleted ORF was PCR-amplified by using an appropriate primer set and pT7-HRS as a template. The resulting fragments were ligated into pCR-Blunt II TOPO and subcloned into pT7-Mock. To obtain pGBKT7-ARMc8α (amino acids 268–648), the deleted ORF was PCR-amplified by using an appropriate primer set and the pcDNA3.1-ARMc8α (previously described [10]) as a template. The resulting fragment was subcloned into the pGBKT7-Mock (BD Biosciences, San Jose, CA) to generate fusion products with the Gal4 activation domain. To construct pGBKT7-ARMc8α and pGBKT7-ARMc8β, pCR-ARMc8α and pCR-ARMcβ (previously described [10]) were digested with EcoRI and BglII , and the resulting fragments were subcloned into pGBKT7-Mock. pT7-HRSΔUIM, which coded for the HRS sequence lacking amino acids 245 to 290, was constructed by the method of site-directed mutagenesis [24]. The expression vector pT7-HRS (S270E) (in which the 270th amino acid serine is substituted glutamic acid for) was constructed with a Quick Change kit (Stratagene, La Jolla, CA). pFLAG-ARMc8α and pEGFP-ARMc8α were constructed previously [10].
Mammalian Cell Culture and Transfection
HEK293 cells were cultured in minimum essential medium. Chinese hamster ovary (CHO) cells were cultured in Ham’s F-12. All media were supplemented with 10% fetal bovine serum, penicillin 100 units/ml, and streptomycin sulfate 100 µg/ml. Plasmid transfections were performed with FuGENE 6 Transfection Reagent (Roche Diagnostics).
RESULTS
Isolation of HRS
To clarify the function of ARMc8, we used a yeast two-hybrid screening assay to search for proteins that may interact with ARMc8. We used the AH109/Y187 yeast strains and construct pGBKT7-ARMc8α (amino acid; 268–648) as a bait to screen 1.0 × 107 independent clones from a human skeletal muscle cDNA library. We isolated 100 clones as candidates. Among them, clone #178 encoded a sequence corresponding to RanBPM, which we previously characterized as being associated with ARMc8 [10]. Sequencing analysis on these candidates revealed that two different clones (clones #87 and #140) coded for sequences corresponding to HRS.
ARMc8α Interacts with HRS In Vivo and Partly Co-Localizes with HRS
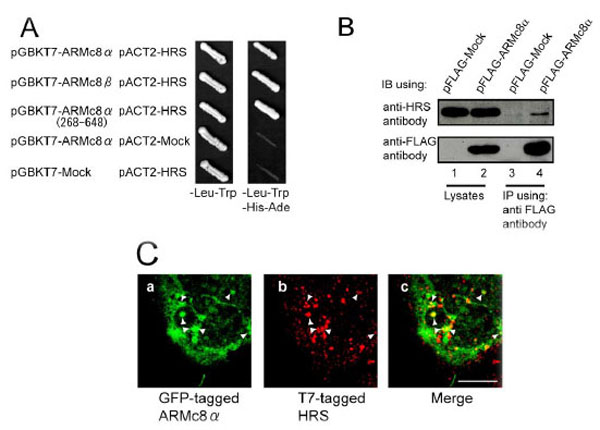
A. AH109 yeast strains were transformed with bait constructs (pGBKT7-ARMc8α, pGBKT7-ARMc8β, pGBKT7-ARMc8α (amino acids 268–648) or pGBKT7-Mock) and prey constructs (pACT2-HRS or pACT2-Mock), and the transformants were inoculated onto a plate containing SD medium lacking leucine and tryptophan to select positive clones transformed with both constructs (bait and prey) (left panel, - Leu-Trp). Colonies positive for growth were restreaked onto a plate of SD medium lacking adenine, histidine, leucine, and tryptophan to select positive clones expressing the reporter genes ADE and HIS3 (right panel, -Ade-His-Leu-Trp).
B. CHO cells were transfected with pFLAG-ARMc8α or pFLAG-Mock, and the cell extracts were immunoprecipitated with an anti-FLAG antibody. The lysates and immunoprecipitates were analyzed by SDS-PAGE, followed by immunoblotting with an anti-FLAG antibody or an anti-HRS antibody. IB, immunoblot. IP, immunoprecipitation.
C. HEK293 cells were co-transfected with pEGFP-ARMc8α and pT7-HRS. Cells were immunostained with a T7•tag antibody and analyzed by confocal microscopy, as described in Material and Methods. GFP-tagged ARMc8α was distributed within the cytoplasm (panel a), as was T7-tagged HRS (panel b). Panel c is a merged image of the two exogenous proteins. Arrowheads indicate typical co-localization. Scale bar represents 10 µm.
We constructed the full-length HRS expression vector pACT2-HRS and confirmed the interaction of ARMc8α, β with HRS by a yeast two-hybrid assay using AH109. ARMc8α was specifically associated with full-length HRS (Fig. 1A the top row) but not with the GAL4-activation domain (Fig. 1A the fourth row from the top). The C-terminal-truncated-ARMc8α (amino acids 268-648) and ARMc8β was also associated with full-length HRS (Fig. 1A the second and third rows from the top). As another negative control, full-length HRS was also not associated with the GAL4-binding domain (Fig. 1A the bottom row). To confirm the association in vivo in mammalian cells, we performed an immunoprecipitation assay. CHO cells were transfected with pFLAG-ARMc8α (Fig. 1B, lane 2, lower panel). After 48 h incubation the exogenous FLAG-tagged ARMc8α was efficiently immunoprecipitated with an anti-FLAG antibody (lane 4, lower panel). Endogenous HRS was co-precipitated with the Flag-tagged ARMc8α (lane 4, upper panel). As a negative control, pFLAG-Mock plasmid was employed in this assay (lanes 1 and 3). These results suggested that ARMc8α associates with HRS in vivo in mammalian cells. To confirm the co-localization of ARMc8α and the early endosomal protein HRS in mammalian cells, we performed an immunofluorescence assay. HEK293 cells were co-transfected with pEGFP-ARMc8α and pT7-HRS, and the cellular distributions of each exogenous protein were examined by confocal immunofluorescence microscopy. The GFP-tagged ARMc8α had a cytoplasmic distribution and in some parts formed small particles (arrowheads in Fig. 1C, panel a). An endosomal distribution of T7-tagged HRS was confirmed (arrowheads in Fig. 1C, panel b). The merged image indicated co-localization of HRS and part of ARMc8α in the endosome (arrowheads in Fig. 1C, panel c). Previous reports have revealed the co-localization of HRS with EEA1, another early endosome marker [25]. The co-localization of exogenous GFP-tagged ARMc8α with endogenous EEA1 was also detected (data not shown). These findings indicated that ARMc8α is partly located on the early endosome. ARMc8α might be involved in membrane trafficking in cooperation with HRS through the early endosome.
Binding Characterization of HRS and ARMc8α
HRS possesses several structural domains that have been characterized [26-28] (Fig. 2A, upper). To identify the binding region of HRS to ARMc8α, we constructed T7-tagged deletion constructs (Fig. 2A, lower), transformed the CHO cells with each deletion construct and FLAG-ARMc8α (Fig. 2B, middle panel, lanes 2 to 7), and then performed an immunoprecipitation assay with an anti-FLAG antibody. The exogenous FLAG-tagged ARMc8α was efficiently immunoprecipitated with an anti-FLAG antibody (Fig. 2B, lanes 9 to 14). As a negative control, we transfected CHO cells with pT7-HRS and pFLAG-Mock plasmid (Fig. 2B, lanes 1, 8). The N-terminal and C-terminal deletion mutants, which possess a proline-rich domain (PRD), co-precipitated with FLAG-tagged ARMc8α (Fig. 2B, lanes 9, 10, 13, and 14). In contrast, the PRD-deletion mutants HRS (amino acids 1–245) and HRS (amino acids 391–777) showed no apparent binding to ARMc8α (lanes 11 and 12). We thought that the band of HRS (amino acids 391–777) might overlap with the band of the mouse IgG heavy chain. We therefore performed an immunoblotting assay using an anti-Omni–probe (rabbit polyclonal antibody), which does not blot mouse IgG (Fig. 2C). HRS (amino acids 335–777) was co-precipitated with FLAG-tagged ARMc8α (Fig. 2C, lane 7), but HRS (amino acids 391–777) again was not (Fig. 2C, lane 8). Furthermore, as described below, lack of the UIM had little or no effect on the interaction (Fig. 3A, lane 2, 3, 4). These results indicate that the proline-rich domain on HRS (amino acids 335–391) is critical for interaction with ARMc8α.
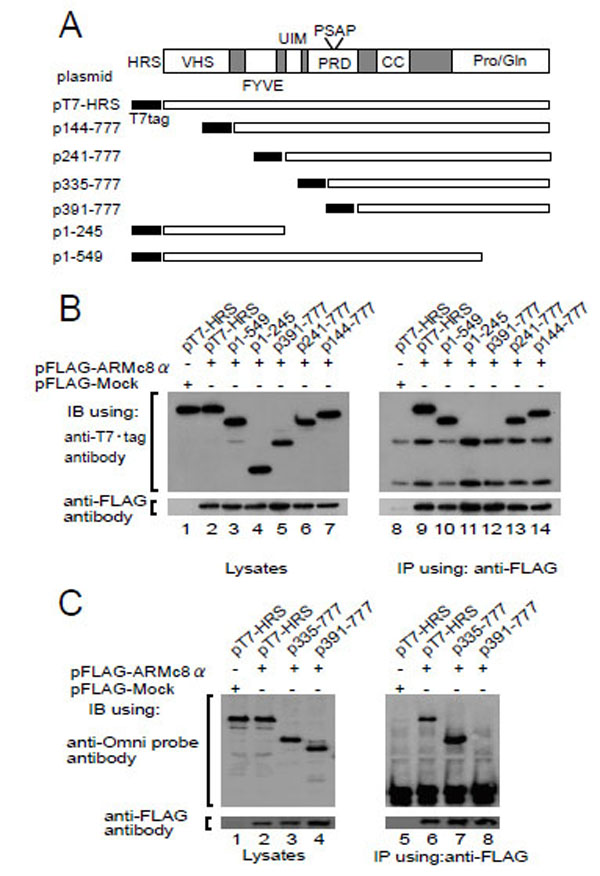
A. upper panel shows the HRS domain organization. Wild type and deletion HRS constructs are shown schematically in the lower panel. Abbreviations are as follows. VHS: Vps27-HRS-STAM; FYVE: Fab1-YOTB-Vac1-EA1; UIM: ubiquitin interacting motif; PRD: prolinerich domain; CC: coiled coil; PSAP: proline, serine, alanine, and proline motif; Pro/Gln: proline-and glutamine-rich domain. B, C. pFLAGARMc8α was co-transfected with each HRS deletion construct in CHO cells. Exogenous ARMc8α was immunoprecipitated with an anti-FLAG antibody. Co-immunoprecipitation of each deleted HRS was analyzed by immunoblotting with an anti-T7•tag antibody (B) and anti-Omni probe (C). As a negative control, cells were co-transfected with pFLAG-Mock and pT7-HRS.
Ubiquitinated Protein Binding to HRS does not Affect the Association Between HRS and ARMc8α
HRS plays essential roles in recognizing monoubiquitinated proteins via the UIM and regulates the endosome/lysosome-dependent degradation of membrane proteins. To examine whether the UIM domain is responsible for the HRS interaction with ARMc8α, we constructed pT7-HRSΔUIM (HRS construct lacking the UIM sequence) and pT7-HRS (S270E) (HRS construct in which the 270th amino acid serine was replaced with glutamic acid; the serine is crucial for the ubiquitin interaction). pFLAG-ARMc8α was co-transfected with pT7-HRS, pT7-HRS (S270E), or pT7-HRSΔUIM in CHO cells (Fig. 3A, lanes 2, 3, and 4, respectively), and then the cell extracts were subjected to an immunoprecipitation assay with an anti-FLAG antibody. The exogenous FLAG-tagged ARMc8α was efficiently immunoprecipitated with an anti-FLAG antibody (Fig. 3A, lanes 2 to 4). As a negative control, pFLAG-Mock plasmid was employed in this assay (Fig. 3A, lane 1). The exogenous HRS, HRSΔUIM, and HRS (S270E) were equally co-precipitated with FLAG-tagged ARMc8α (Fig. 3A, lanes 2 to 4). Monoubiquitinated proteins were also co-precipitated with HRS (Fig. 3A, bottom panel, lane 2), but faintly with HRS (S270E) (Fig. 3A, lane 3) and HRSΔUIM (Fig. 3A, lane 4). UIM containing proteins interact with ubiquitinated proteins. Generally UIM binds to both monoubiquitin and polyubiquitin chains. But there are different tendencies whether to bind to monoubiquitin or polyubiquitin chains in each UIM containing proteins. The UIM of HRS tends to bind to monoubiquitin more strongly than any other UIM containing proteins do [29]. So the ubiquitinated proteins immunoprecipitated with HRS are supoosed to be mainly monoubiquitinated proteins. These results indicated that monoubiquitinated protein binding to HRS does not affect the association between HRS and ARMc8α.
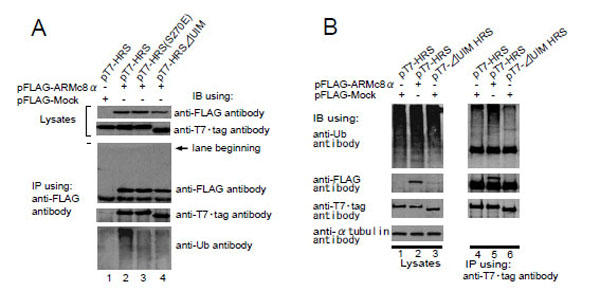
A. In CHO cells, pFLAG-ARMc8α was co-transfected with pT7-HRS, pT7-HRS (S270E), or pT7-HRSΔUIIM, as indicated (lanes 2 to 4). Exogenous ARMc8α was immunoprecipitated with anti-FLAG antibody. Co-immunoprecipitations of each deleted HRS and ubiquitin were analyzed by immunoblotting with the antibodies indicated. As a negative control, cells were co-transfected with pFLAG-Mock and pT7-HRS (lane 1). Arrow indicates the location of the lane beginning.
B. In CHO cells, pFLAG-ARMc8α or pFLAG-Mock was co-transfected with pT7-HRS or pT7-HRSΔUIM, as indicated (lanes 1 to 3). The transfected cells were lysed and the exogenous HRS and each deleted HRS were immunoprecipitated with T7•tag antibody. This was followed by immunoblotting with the antibodies shown (lanes 4 to 6). IB, immunoblot. IP, immunoprecipitation.
Exogenously Expressed ARMc8α Increases the Interaction of Ubiquitinated Protein with Exogenously Expressed HRS Via the UIM Domain
We next examined the possibility that ARMc8α affected the binding of monoubiquitinated protein to the UIM of HRS. CHO cells were transfected with pT7-HRS or pT7-HRSΔUIM and subjected to an immunoprecipitation assay. The exogenous T7-tagged HRS and HRSΔUIM were efficiently immunoprecipitated with T7·Tag monoclonal antibody (Fig. 3B, third panel from the top, lanes 4 to 6). Monoubiquitinated proteins were co-precipitated with the T7-tagged HRS (Fig. 3B, lane 4). The UIM is essential for monoubiquitinated protein binding to HRS, because the co-precipitation with HRSΔUIM was faint (Fig. 3B, lane 6). When pFLAG-ARMc8α was co-transfected in CHO cells with pT7-HRS, the overexpressed ARMc8α resulted in increased monoubiquitinated protein binding to the T7-tagged HRS (lane 5), suggesting that ARMc8α upregulates the binding of monoubiquitinated proteins to the UIM. Considering the fact that HRS regulates various cellular processes via UIM interaction with monoubiquitinated proteins, this finding is very interesting and seems to support the hypothesis that, by interacting with HRS, ARMc8α partly mediates the association of HRS to monoubiquitinated proteins.
DISCUSSION
We found that HRS binds to ARMc8α via the PRD, which has been characterized as a site of protein interaction with tumor suppressor gene 101 (TSG101) [28], EGFR pathway substrate clone 15 and Sorting nexin 1 [25]. Indeed, we demonstrated that the PRD (HRS amino acids 335-391) is a critical binding site for ARMc8α interaction. Since the PSAP motif, which is crucially implicated in HRS interaction with TSG101 [28], exited at the N-terminal sequence of PRD (HRS amino acids 348-351) as a binding region, the PSAP motif may play an essential role in HRS- ARMc8α interaction.
Although we have not yet revealed the mechanism by which overexpressed ARMc8α upregulates the binding of monoubiquitinated proteins to HRS, we can hypothesize that ARMc8α competitively obstructs the binding of TSG101 to the PSAP motif in HRS; since TSG101 is also essential for the sorting of monoubiquitinated proteins into the MVB, overexpressed ARMc8α might thus increase the binding of undegraded, monoubiquitinated proteins. Actually to clarify the function of ARMc8α, we also have been exploring the effect of siRNA-mediated gene specific silencing of ARMc8α on the interaction between UIM of HRS and monoubiquitinated proteins. However, so far we haven’t yet found a significant effect.
On the basis of this finding we can reach a hypothesis that ARMc8α has high potency and only a small amount is required to mediate the interaction of HRS with monoubiquitinated proteins. In other words, the amount of ARMc8α downregulated by siRNA-mediated gene silencing might still be enough to normally regulate the interaction of HRS with monoubiquitinated proteins while overexpression of ARMc8α might function suppressively as dominant-negative or, as described above, competitively obstruct the binding of TSG101 to the HRS leading to stagnated HRS-mediated lysosomal trafficking and resulting in increasing the undegraded ubiquitinated proteins which could associate with HRS. Nevertheless, we believe that much more optimized experimental model where gene downregulation level is much more rigorously controlled would be required to draw a conclusion.
HRS plays an essential role in the endosome/lysosome-dependent degradation of receptor proteins via endocytosis. HRS associates with monoubiquitinated receptor proteins through the UIM and sorts them into the MVB. EGFR is a typical molecule downregulated by this mechanism [18]. In our experiment we found that ARMc8α co-localizes with HRS and EEA1 and that overexpressed ARMc8α stimulates the binding of monoubiquitinated proteins to HRS. Therefore, we supposed that overexpressed or downregulated ARMc8α might affect EGFR degradation via HRS. Thus, to estimate the effect of ARMc8α , we examined EGFR degradation after stimulation of epidermal growth factor (EGF), which is a ligand to induce EGFR downregulation [18], in cells transiently transfected with ARMc8α expression vector or gene-silenced by siRNA with ARMc8α specific primer. Also we established stable ARMc8α -overexpressing and shRNA-mediated knockdown cell line on the same purpose. However, we couldn’t detect a significant effect of ARMc8α in these experiments. Moreover EGF stimulation didn’t show any effects on the association of HRS with ARMc8α in both immunoprecipitation assay and immunofluorescence microscopy (data not shown). So, taken together, we have speculated that ARMc8α might not regulate EGFR degradation in cooperation with HRS.
Although we couldn’t find any effects of ARMc8α on EGFR degradation, identifying the monoubiquitinated protein that is degraded in an ARMc8α -dependent manner will be very important. Because many receptor proteins, such as Drosophila signaling receptors [19], Toll-like receptor 4 [21], and VEGFR2 [20] have been characterized as molecules down-regulated by an HRS-dependent mechanism, these receptors are candidates. Furthermore, HRS is also known to play critical roles in the recycling of certain receptor proteins such as transferrin receptor and β2-adrenergic receptor [30]; the autophagic degradation pathway [31]; and stabilization and degradation of STAM (signal transducting adaptor molecule) proteins [32]. The cellular transport and degradation of several ubiquitinated proteins might be regulated by HRS in cooperation with ARMc8α, although the details of this mechanism are unclear.
Here, we analyzed the interaction of ARMc8α with HRS in mammalian cells. HRS is well conserved through eukaryotes as a key component regulating endosomal sorting. In yeasts, as well as in mammalian cells, ubiquitination plays critical roles in both proteasome- and vacuole-dependent protein degradation, and ubiquitination functions as a signal for entry to the lysosome degradation pathway [18]. As mentioned in the Introduction, ARMc8α is human homolog of yeast vid28. It would be interesting to analyze the possible association of vid28 with vps27, the yeast homolog of HRS. This investigation of vid28 and vps27 might help to clarify the function of ARMc8α and CTLH complex in mammalian cells, because the functions of these yeast homologs have been partly characterized.
ACKNOWLEDGEMENT
This work was supported in part by grant to A. Ueda from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]
[PubMed Link]
[PubMed Link] [PMC Link]


