All published articles of this journal are available on ScienceDirect.
Warfarin Dose Maintenance Associated with CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) Gene Polymorphism in North Coastal Andhra Pradesh
Abstract
Background
Deep Vein Thrombosis (DVT) is a high-risk condition that necessitates the use of oral anticoagulants for treatment. Warfarin, a common anticoagulant, exhibits varying levels of efficacy and toxicity among individuals. The CYP2C9 gene promoter polymorphism significantly influences the dosage requirements, a factor that remains underexplored in the contemporary Indian population.
Objectives
This study aimed to investigate the influence of CYP2C9 gene polymorphisms on warfarin dosage due to pharmacogenetic effects. Specifically, it examined the prevalence of the CYP2C9 polymorphic alleles *2 and *3 and their correlation with warfarin dosage in the South Indian Population (NCAP).
Methods
The study involved 96 warfarin-treated patients to determine the genotype frequency of common CYP2C9 polymorphisms. The genotypes of CYP2C92 and CYP2C93 polymorphisms were analyzed using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. A one-way analysis of variance (ANOVA) was conducted to ascertain dosage variation across genotypes.
Results
The study found that the frequencies of the two variations were 25.5% for CYP2C92 and 40.6% for CYP2C93. Patients with a homozygous wild-type genotype for CYP2C9 (*1/*1) required a daily warfarin dose of 4.07 ± 1.75 mg, significantly higher than the *1/*2, *1/*3 (2.93 ± 2.03 mg, p <0.0001) and *2/*2, *2/*3, and *3/*3 patients (1.54± 1.05 mg, p = 0.002). The study also revealed a distinct allelic frequency of CYP2C9 polymorphisms in the study population compared to other populations.
Conclusion
Given the relatively high prevalence of CYP2C9 polymorphisms in the studied population, practitioners should consider these findings to minimize the risk of bleeding when prescribing warfarin.
1. INTRODUCTION
Warfarin is an anticoagulant that is used to treat and prevent issues with arterial and venous thrombosis. Additionally, warfarin is given to prevent pulmonary embolism, stroke, and transient ischemic attack [1, 2]. It has a narrow therapeutic index and requires frequent monitoring of blood prothrombin time to decrease the risk of bleeding [3]. The dose requirement is estimated by measurement of PT- INR tests. Regular estimation of the coagulation rate is essential for any fixed dose. Every patient's daily dosage is modified in order to keep their affected individual's INR within the targeted range [4]. The variation in the dosage response of the anticoagulant effect is observed in large inter-individual, intra-individual, and inter-ethnic patients [5]. There are no fixed doses of warfarin therapy, and dose requirements are encouraged by weight, age, medical condition, concomitant use of other pills, and genetic factors [6]. All of these factors must be taken into account to improve the profile of benefits associated with warfarin therapy [7].
CYP2C9 and VKORC1 are key enzymes in the pharmacokinetic and pharmacodynamic routes of warfarin [8-10]. Vitamin K epoxide reductase complex, subunit 1 (VKORC1), is the site of a target for warfarin activity, which inhibits the anticoagulant effect [11, 12]. The metabolism of warfarin is carried out by the CYP2C9 enzyme, and warfarin has R and S racemic mixtures. S warfarin is more potent than R warfarin [11]. The CYP2C9 enzyme transforms S-warfarin into its active metabolite [13]. SNPs in CYP2C9, such as * 2 (rs1799853) and * 3 (rs1057910), are frequently found and have a considerable impact on the dose of warfarin [14, 15]. They account for between 9.6% and 20.6% of variations in individualized medicine [16]. The wild-type variants of the gene have a reduced metabolic capacity for warfarin [17]. The variants of CYP2C9 contribute as a genetic factor along with demographic factors to warfarin dose variability. It has been observed that if the level of the drug within the blood is much lower, healing objectives cannot be accomplished, and there is a threat of blood clots. However, if it is too high, hemorrhage is more likely to occur [18]. By genotyping Single Nucleotide Polymorphisms (SNPs) in genes that regulate warfarin metabolism or its targeted enzymes, it is feasible to establish the ideal therapeutic dose of warfarin [19]. Warfarin is metabolized by the enzyme CYP2C9, and a number of frequent variations have been found. Among these variants, CYP2C9*2 rs1799853 (c.430 C>T, p. Arg144Cys) and CYP2C9*3 rs1057910 (c.1075 A>C, p.Ileu359Leu) are the most common functional ones [20]. The encoding of SNPs affects the enzymatic activity and metabolic capacity of the enzyme CYP2C9 by substituting amino acids. Patients with one or two of these variations require less warfarin and are more likely to bleed than patients with the wild-type gene.
In this study, we examined the genotypes and alleles of CYP2C9 and discovered that individuals with deep vein thrombosis frequently carry a few minor alleles. We also found that some polymorphic variants of CYP2C9 can affect how much of a drug a person needs to achieve the desired effect.
2. MATERIALS AND METHODS
The study was conducted in Gayatri Vidya Parishad Institute of Health Care and Medical Technology, Visakhapatnam. The study was started after receiving ethical approval from the Institutional Ethics Committee. Informed consent was obtained from all participants in either their native language or in English. The institutional ethics committee approved the study in October, 2020, with IEC GVPIHCMT/IEC/20201019/01.
2.1. Aim of the Study
This study aimed to study the variability in the alleles of CYP2C9 and compare the effect of “alleles of CYP2C9*2” and “CYP2C9*3” on the dose of warfarin.
2.2. Study Population
This is a pragmatic randomized study on 96 patients, comprising 45 males and 51 females with DVT, who have been on warfarin therapy for at least three months with a targeted INR range of 2.5-3.5. The sample size was calculated according to the Journal of Genetic Engineering and Biotechnology Research [21].
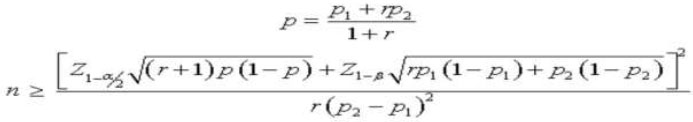 |
Alpha (α) - 0.05, Beta (β) – 0.2
Proportion in group 1 – 0.13
Proportion in group 2 – 0.3
Minimum sample size needed for group 1: 96
Minimum sample size needed for group 2: 96
| Patient Characteristics | Test Group |
|---|---|
| Sex | - |
| Male | 45 (47%) |
| Female | 51 (53%) |
| Age | - |
| Mean | 46 |
| Median | 47 |
| Range | 21-66 |
| Weight | - |
| Mean | 47 |
| Median | 45.5 |
| Range | 33-75 |
This study included deep vein thrombotic patients aged between 18 and 65 years who were stable on warfarin maintenance doses. Participants who did not give consent and those who had risk factors, such as hypertension, liver diseases, or diabetes mellitus, using CYP2C9 inducers and inhibitors were excluded from this study. Table 1 lists the characteristics of the patients.
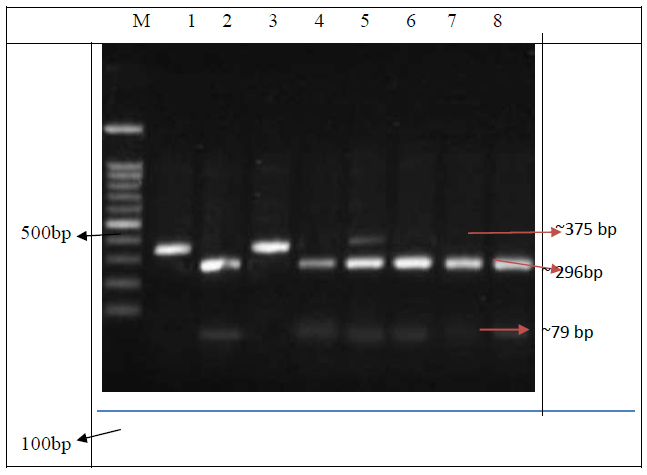
Restriction digested banding patterns of CYP2C9*2 gene. Lane M, 100-bp ladder; Lanes 1 and 3 were TT homozygous mutants with a fragment of 375-bp; Lanes 2, 4, 6, 7, and 8 were CC wild homozygous with fragments of 296-bp and79-bp. Lane 5 was CT heterozygous with fragments of 375- bp, 296-bp, and 79-bp.
2.3. Statistical Analysis
The data was analyzed using the institutional SPSS program. The data are represented as frequencies and percentages, and the variables are shown as mean and median when suitable. The ANOVA test was performed to assess the statistically significant variations in the daily maintenance dose of warfarin among the various genotype groups. For each variable, odds ratios were determined. A statistically significant p-value was defined as < 0.05.
2.4. Sampling and DNA Extraction
Each participant provided two milliliters of blood, which was then placed into tubes containing ethylenedi- aminetetraacetic acid (EDTA) to avoid coagulation. After running these samples for 15 minutes at 40°C at 4000 rpm, the plasma was separated, and a PT-INR test was performed. Within an hour of the blood sample being taken, this test was conducted. For DNA analysis, the pellet was stored at -20°C. DNA was extracted using the salting out method. After DNA extraction, PCR-RFLP was performed.
2.5. Genotyping of CYP2C9 Allele
The CYP2C9*2 and CYP2C9*3 polymorphisms were found by using a PCR-RFLP method. The CYP2C9*2 (5'-CACTGGCTGAAAGAGCTAACAGAG-3' and 5'GTGATATGG AGTAGGGTCACCCAC- 3' amplified 375 bp and the CYP2C9*3 (5'- AGGAAGAGATTGAACGTGTGA -3' and 5'- GGCAGGCTGGTGGGGGAAGGCCAA - 3' amplified 130 bp. We executed the PCR reactions as described by Sajja et al. (2004). PCR cyclic conditions consisted of the following steps: an initial denaturing of 5 minutes at 95°C, followed by 35 denaturing cycles at 950C for 30 s, annealing at 59°c for 45 s, and extension at 720C for 45 s, with a final extension of 10 minutes at 720C, using the Biorad MJ Mini TM thermocycler. The digestion products were examined on a 2% agarose gel.
3. RESULTS
Gene amplification was confirmed by electrophoresis. PCR was used to amplify a 375bp DNA fragment for CYP2C9*2. A single band at 375 bp indicated the presence of the mutant allele T. The C allele was digested into two fragments of 296 bp and 79 bp (Fig. 1). The banding pattern of the A allele for CYP2C9 *3 was 130 bp, while the banding pattern of the C allele was 104 bp and 26 bp (Fig. 2).
The CYP2C9*2 genotype frequencies for the CC, CT, and TT genotypes were 64.6%, 19.8%, and 15.6%, respectively. The CYP2C9*3 genotype frequencies for the AA, AC, and CC genotypes were 39.6%, 39.6%, and 20.8%, respectively. The mutant allele for CYP2C9*2 (T allele) was determined to be 25.5% of the 96 patients studied, with 19 being heterozygous (CT; 19.8%) and 15 being homozygous (TT; 15.6%). The mutant allele for CYP2C9*3 (Callele) was determined to be 40.6% common. There were 38 heterozygous people (AC; 39.6%) and twenty homozygous people (CC; 20.8%). The genotype frequencies of wild and variant alleles were 28 (29.2%), 06 (6.3%), 23 (24%),04 (4.2%), 25 (26%), and 10 (10.4%) for *1/*1, *1/*2, *1/*3, *2/*2, *2/*3, and *3/*3, respectively (Table 2).
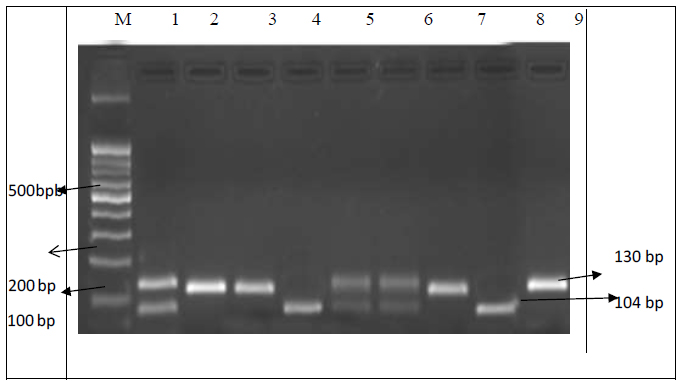
Restriction digested banding patterns of CYP2C9*3 gene. Lane M, 100-bp ladder; Lanes 1, 5, 6, and 12 were CA heterozygous with fragments of 130-bp, 110-bp, and 26-bp; Lanes 2, 3, 7, 9, and 5 were wild type with a fragment of 130-bp; Lanes 4 and 8 were homozygous mutant CC with fragments of 130-bp and 26-bp.
| CYP2C9 | All Subjects (n:96) | Males (n:45) | Females (n:51) | p-Value | Odds ratio |
|---|---|---|---|---|---|
| *1/*1 | 28(29.2) | 13 (28.8) | 15 (29.5) | 0.866 | 0.98 (0.4-2.36) |
| *1/*2 | 06(6.3) | 3 (6.7) | 3 (5.8) | 0.791 | 1.14 (0.22-5.97) |
| *1/*3 | 23 (24.0) | 10 (22.3) | 13 (25.5) | 0.892 | 0.84 (0.33-2.15) |
| *2/*2 | 04 (4.2) | 2 (4.5) | 2 (3.9) | 0.701 | 1.14 (0.15-8.44) |
| *2/*3 | 25 (26.0) | 11(24.4) | 14 (27.5) | 0.918 | 0.86 (0.34-2.14 |
| *3/*3 | 10 (10.4) | 6 (13.3) | 4 (7.8) | 0.586 | 1.81 (0.48-6.87) |
Patients with the homozygous wild-type CYP2C9 (*1/*1) genotype received a daily mean dosage of warfarin (4.07± 1.75 mg), according to a comparison of warfarin daily maintenance doses between genotype groups. It was significantly higher than in patients with *1/*2, *1/*3 (2.93 ± 2.03 mg, p<0.001) and *2/*2, *2/*3, and *3/*3 (1.54 ± 1.05 mg, p = 0.002). In individuals with wild-type CYP2C9*1, the mean daily warfarin dose was noticeably higher. Furthermore, the mean daily warfarin dose differed statistically between heterozygous and homozygous genotypes.
4. DISCUSSION
DVT is most often caused by factors, such as age, hospitalization, pregnancy, hormone therapy, cancer treatment, and surgery. These conditions can also increase a person's risk of developing DVT due to genetic variations in genes that control drug transport and metabolism [22]. Several studies have found that genetic factors account for more than 60% of the risk of DVT [23, 24]. Each individual requires a different dosage of the anticoagulant warfarin, which is metabolized by CYP2C9. The anticoagulant dosage requirements are determined by varying allele combinations in CYP2C9 and VKORC1, which affect the medication clearance rate [22]. Individualized warfarin dosages would lead to fewer adverse effects and decrease hospital and patient healthcare expenses [25].
Patients with CYP2C9*2 and CYP2C9*3 alleles have longer half-lives for S-warfarin, which is more bioactive, thus resulting in a lower dose requirement, a higher risk of bleeding, and take potentially more time to achieve steady-state compared to those with the wild-type genotype [26]. For CYP2C9*2 and CYP2C9*3 in several racial groupings, differing allelic frequencies have been recorded.
It is critical to investigate the pattern of genetic variation distribution in various parts of the country to develop an appropriate warfarin dosing algorithm and provide better information on the affiliation among CYP2C9 versions and sensitivity to warfarin. The current study examined the frequency of CYP2C9 alleles and polymorphisms (rs1799853, rs1057910) in 96 DVT patients from South India. The CYP2C9 allele *1 was found in 33.9% of the patients in our study, while variant alleles 2 and 3 were found in 25.5% and 40.6% of the patients, respectively. In different populations, CYP2C9*2 and CYP2C9*3 allelic frequencies were compared (Iranian, Turkish, Indian, Chinese, Malaysian, and European). When compared to our study population, the frequency of CYP2C9*2 and CYP2C9*3 variants (9.1% and 10%, respectively) was lower in the northeast of Iran [27]. In contrast to the Gujarati Indians with 4.90% and 3.92% [28], 2.6% and 4.6% of Chinese [29], and 12.5% and 8.5% of the Europeans [30], the CYP2C9*2 and CYP2C9*3 frequencies in our study were different. Furthermore, the frequency of CYP2C9*2 allele genotypes differed between South and North Indians, which are ethno-geographically distinct populations [31]. The CYP2C9*3 allele was found to be more common in this population than in other Asian people [32].
The impact of CYP2C9 polymorphism on the daily dose of warfarin was also examined. The natural allele *1 requires more warfarin than patients with CYP2C9 mutant alleles *2 and *3. There was no statistically significant difference between homozygous and heterozygous mutant individuals, whereas individuals with homozygous wild-type *1/*1 genotype needed a notably higher dose. The first documented relationship between CYP2C9 polymorphisms and warfarin dosage requirements was discovered in 1999 [33]. Several studies have demonstrated that CYP2C9 allele variations are linked to a lower average dose of warfarin than wild-type alleles [34]. Several clinical trials have also been conducted using genotype-guided assays, such as CYP2C9 genotyping. Clinical trials demonstrated that the use of genotype-guided treatments, including genotyping for CYP2C9 in dosing, was effective. According to the Clarification of Optimal Anticoagulation through Genetics (COAG) by Kimmel et al., genotype-guided warfarin dose did not enhance anticoagulation control based on time spent within the therapeutic range during the first four weeks of treatment [35].
It has been demonstrated that the patient should complete at least 10-12 weeks of initial therapy. Moreover, the European Pharmacogenetics of Anticoagulant Medication (EU-PACT) warfarin trial found that genotype-based dosing on days 1-5 was superior to conventional therapy in terms of time spent within the desired therapeutic range over three months [36]. In recently published data, the Genetic Informatics Trial (GIFT) research revealed that genotype-directed dosage produced better results for patients than controls who received doses determined by a clinically guided algorithm.
CONCLUSION
The present study reported the high prevalence of CYP2C9 polymorphisms. CYP2C9 is a gene that codes for an enzyme involved in the metabolism of warfarin, a commonly prescribed anticoagulant. Polymorphisms in the CYP2C9 gene can lead to variations in the activity of enzymes, affecting how individuals metabolize warfarin. This implies that clinicians should be cautious and take into consideration the genetic variability in CYP2C9 when administering warfarin to patients. Since warfarin is used to prevent blood clotting, understanding the patient's genetic makeup can be crucial for determining the appropriate dosage. Individuals with certain CYP2C9 polymorphisms may metabolize warfarin more slowly or rapidly, affecting its efficacy and increasing the risk of bleeding or clotting events.
Clinicians might consider genetic testing for CYP2C9 variants before prescribing warfarin to tailor the dosage to an individual's specific genetic profile. This personalized approach, known as pharmacogenomics, can help optimize treatment outcomes and minimize adverse effects. It underscores the importance of incorporating genetic information into clinical decision-making, particularly in the case of drugs with a narrow therapeutic index like warfarin. The significance of integrating genetic data into clinical decision-making needs to be emphasized, especially for medications like warfarin that have a narrow therapeutic index.
AUTHORS’ CONTRIBUTION
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| DVT | = Deep Vein Thrombosis |
| PE | = Pulmonary Embolism |
| CYP | = Cytochrome |
| VKOR | = Vitamin K epoxide reductase |
| PCR | = Polymerase Chain Reaction |
| dNTPs | = Deoxynucleoside triphosphates |
| Bp | = Base Pair |
| EDTA | = Ethidum diaminetetraacetic acid |
| TE | = Tris EDTA |
| RFLP | = Restriction Fragment Length Polymorphism |
| PT | = Prothrombin Time |
| INR | = International Normalized Ratio |
| EM | = Extensive Metaboliser |
| IM | = Intermediate Metaboliser |
| PM | = Poor Metabolizer |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was conducted at Gayatri Vidya Parishad Institute of Health Care and Medical Technology, Visakhapatnam. It was started after receiving ethical approval from the Institutional Ethics Committee with IEC GVPIHCMT/IEC/20201019/01.
HUMAN AND ANIMAL RIGHTS
All procedures performed in studies involving human participants were in accordance with the ethical standards of institutional and/or research committee and with the 1975 Declaration of Helsinki, as revised in 2013.
CONSENT FOR PUBLICATION
Informed consent was obtained from all participants in either their native language or in English.


