All published articles of this journal are available on ScienceDirect.
Prevalence of Extended-Spectrum β-lactamases in Enterobacteriaceae Isolated from Polluted Wild Fish
Abstract
Background:
Antibiotic resistance is becoming a major public health concern worldwide. In marine animals, pollution is associated with the emergence of extended-spectrum β-lactamase (ESBL)-expressing bacteria, resulting in antibiotic resistance. However, the prevalence of these bacteria in wild fish has not been reported.
Objective:
Accordingly, in this study, we explored the influence of pollution oxidative stress on the incidence of ESBL-expressing Enterobacteriaceae in the gut of wild fish species from the Red Sea coastal region of Jeddah City, Saudi Arabia. Additionally, we evaluated the incidence of varied ESBL genes contributing to the ESBL+ phenotype.
Methods:
Antibiotic susceptibility tests were performed using cephalosporins and carbapenems against ESBL- and carbapenem-resistant Enterobacteriaceae (CRE)-producing bacteria. Frequent genes contributing to the ESBL+ phenotype were analyzed. Primers targeting ESBLs (e.g., blaCTX, blaSHV, blaTEM, and blaOXA) were used in polymerase chain reaction assays to detect the ESBL+ phenotype.
Results:
Screening results from the polluted site revealed ESBL-resistant Klebsiella pneumoniae B8 and CRE-resistant Morganella morganii A4. The evolution of the blaCTX-M gene in M. morganii was a consequence of aquatic pollution. The other isolates Acinetobacter pittii and Providencia rettgeri were found in the clean reference site. The isolate M. morganii showed resistance to most mutual antibiotics and expressed some β-lactamase genes.
Conclusion:
Our findings provide useful data for selecting marine molecular genomic biomarkers caused by aquatic pollution.
1. INTRODUCTION
Antimicrobial resistance owing to long-term exposure to certain pollutants is becoming a major public health concern [1]. Numerous genetic mechanisms are involved in antibiotic resistance in bacteria. Extended-spectrum β-lactamase (ESBL) resistance mechanisms are present in most Enterobacteriaceae species worldwide (Goossens and Grabein, 2005; Nicolas-Chanoine et al., 2014).
ESBLs are categorized as CTX-M, TEM, and SHV types [2], and some ESBLs confer resistance to third-generation antibiotics, such as cephalosporins (ceftazidime and cefotaxime), but can be inhibited by the β-lactamase inhibitor clavulanic acid. These enzymes are also unable to hydrolyze carbapenems, such as imipenem [3]. The best antibiotics against ESBL-carrying bacteria are carbapenems; however, Enterobacteriaceae capable of producing resistant β-lactamases have yielded Carbapenem-Resistant Enterobacteriaceae (CRE), and these organisms have spread worldwide [4, 5].
The presence of ESBL-expressing bacteria in seafood can lead to the transmission of resistance-related factors to other clinically important bacteria. For example, Vibrio pathogens are common environmental contaminants found in seafood. Additionally, studies have shown that infected water bodies can cause transmission of other clinically important bacteria, such as Escherichia coli and Salmonella enterica [6]. Seawater is also subjected to continuous anthropogenic pollution; therefore, ESBL-expressing Enterobacteriaceae in seafood should be evaluated.
Accordingly, in this study, we explored the influence of pollution oxidative stress on the incidence of ESBL-expressing Enterobacteriaceae in the gut of wild fish species from the Red Sea coastal region of Jeddah City, Saudi Arabia and examined the incidence of ESBL genes contributing to the ESBL+ phenotype.
2. MATERIALS AND METHODS
2.1. Sampling and Sample Preparation
Random 200 wild fish were sampled from a Polluted (PO) area near the location 21°16′14.2″N, 39°07′22.4″E, where an abundance of sewage was released, and a Clean Reference (CR) area near the location 21°12′24.3″N, 39°09′57.9″E in the Red Sea coastal region of Saudi Arabia. The annual temperature of the seawater in Jeddah city ranged from 25 to 29°C. Only fish over 20 cm in length were sampled. Experimental fish were randomly harvested, placed in clean, sealable plastic bags, transferred to the laboratory on ice, and stored at 4°C for processing.
The fish were dissected on the same day, and the intestines were removed aseptically from the abdominal cavity of each fish. The contents were carefully collected and labeled according to the area of collection. Seawater was collected from three different sites using sterile glass vials. To avoid surface contamination, water samples were collected from 15-20 cm below the surface of the water. Water quality for both sites was assessed to determine pH, Total Dissolved Solids (TDS), Electrical Conductivity (EC), Total Solids (TS), Chemical Oxygen Demand (COD) [7], total suspended solids (Berntsson et al., 2013), Biochemical Oxygen Demand (BOD), and pH. Standard protocols were used to estimate all other parameters [8]. Statistical analysis including mean, standard deviation and test of statistical significance, t-test were performed using MS Excel version 2013.
2.2. Antibiotic Susceptibility of Resistant Strains
Fecal samples of fish were cultured in enrichment medium Lysogeny Broth (LB) at 37°C for 24 h. Resistance to meropenem, imipenem, cefotaxime, and ceftazidime was determined for isolates with potential carriers of CRE or ESBLs. The agar dilution method was used for the assessment of resistant strains [9]. Two inoculums from the culture broth were subcultured on Mac Conkey agar supplemented with 2 μg/mL ceftazidme or cefotaxime [10, 11] for the selection of CRE or with 1 μg/mL imipenem or meropenem [12] for the selection of ESBLs. The agar cultures were incubated at 37°C for 24h. To purify targeted strains, colonies were subcultured on MacConkey medium one more time with a higher antibiotic concentration (4 µg/mL). Growing bacteria with pink pigmentation were subcultured on enrichment LB agar medium. The results were described according to CLSI-2011 procedures, and isolates were assumed resistant to both ceftazidime and cefotaxime if the value of the Minimal Inhibitory Concentration (MIC) was greater than or equal to 2 μg/mL in accordance with CLSI standards [13]. Ten individual isolates were recovered for 16S rRNA and Sanger sequencing.
2.3. DNA Isolation and Genotyping
Bacterial DNA was mined using InstaGene Matrix (Bio-Rad, Hercules, CA, USA; cat. no. 732-6030) accordingly to the manufacturer’s instructions, and the concentration of DNA was measured using Picogreen technique with Victor 3 fluorometry (Invitrogen, Carlsbad, CA, USA; cat. no. P7589). An Agilent Technologies 2100 analyzer with 1000 chip DNA was used to check the distribution of template size in order to verify the size of the Polymerase Chain Reaction (PCR)-enhanced fragment. A portion of each removed DNA sample was used for 16S rDNA magnification. The 16S rDNA was enlarged with universal primers 1492R (5′-TACGGYTACCTTGTTAC GACTT-3′) and 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) (Stackebrandt and Goodfellow, 1991). Twenty nanograms of genomic DNA was subjected to PCR in a 30-μL reaction using Dr. MAX DNA polymerase (Doctor Protein Inc., Korea; cat. no. DR00302). The PCR program was as follows: 35 cycles of 5 min at 95°C, 30 s at 95°C, 30 s at 55°C, 1.5 min at 72°C, and 7 min at 4°C [14]. Subsequently, using a multiscreen filter-plate (Millipore Corp., USA), the extension products were cleaned. The products were then sequenced using a Big Dye PRISM Terminator V 3.1 Kit for Sequence Cycling (Applied Biosystems, Foster City, CA, USA). The extension products were supplemented with Hi-Di formamide (Applied Biosystems), and the mixture was tested with an ABI 3730-XL Prism Genomic analyzer (Applied Biosystems) at 95°C for 5 min, followed by incubation for 5 min on ice. Sequencing of DNA isolates was achieved using Macrogen Inc. (Seoul, Korea), and a phylogenetic tree to compare the ESBL gene carrier strains was constructed using the neighbor-joining method and evaluated using the interior branch test method with Mega 4 software.
2.4. Detection of ESBL Genes by PCR
The inclusive term ESBLs included SHV-1, TEM-1, OXA, and CTX-M type enzymes [17]. These enzymes could hydrolyze and deactivate various beneficial β-lactam antimicrobials [18]. Genes encoding ESBLs (blaCTX, blaSHV, blaTEM, and blaOXA) were distinguished by PCR with definite resistance gene primers, and sequencing was performed using PCR products, as shown in Table 1. The PCR program was as follows: 35 cycles of 5 min at 95°C, 30 s at 95°C, 30 s at 55°C, 1.5 min at 72°C, and 7 min at 4°C [14]. Enhanced PCR fragments were assessed by 2% agarose gel electrophoresis, screened using ultraviolet illumination, and imaged with a gel documentation system (Gel-Doc 2000; Bio-Rad). The sequencing of DNA for antibiotic-resistance genes was performed by Macrogen Inc. (Seoul, Korea).
| No. | Target Gene | Primer Sequence | Amplicon Size (bp) | References | |
| 1 | blaCTX-M | 5′ (For) | CGCTGTTGTTAGGAAGTGTG | 569 | [41] |
| 3′ (Rev) | GGCTGGGTGAAGTAAGTGAC | ||||
| 2 | blaOXA | 5′ (For) | ATATCTCTACTGTTGCATCTCC | 619 | [42] |
| 3′ (Rev) | AAACCCTTCAAACCATCC | ||||
| 3 | blaTEM | 5′ (For) | TGCAACAGTGCCTCTCGATA | 717 | [43] |
| 3′ (Rev) | CTCGTGCACCCAACTGATCT | ||||
| 4 | blaSHV | 5′ (For) | GGTTATGCGTTATATTCGCC | 867 | [44] |
| 3′ (Rev) | GGTTAGCGTTGCCAGTGCTC | ||||
3. RESULTS
3.1. Water Analysis
Water quality data from PO versus CR areas were compared. Both samples were assessed for pH, EC, TS, TDS, BOD, and COD, as summarized in Table 2. The samples from PO areas showed greater pollution than samples from CR areas owing to the higher COD of 65.70 ± 0.53 mg/L (*P ≤ 0.05). The BOD of polluted water was not very high, suggesting that the amount of biodegradable organic pollutant matter was low. The concentration of salt was similar between the two sample areas, indicating that pollution did not affect the salinity of the water. The TS content in samples from PO areas (58.06 ± 0.26 mg/L) was higher than that in samples from CR area (41.8 ± 0.10 mg/L) owing to the presence of additional pollutants in the water (* ≤ 0.05). In PO samples, most of the pollutants were in the dissolved form, and the TS content the fraction of TDS was quite high (56.67 ± 0.06 mg/L), whereas the TSS was exceedingly low (1.49 ± 0.007 mg/L). Pollution did not appear to affect the pH of the water.
| Parameters | PO | CR |
| COD (mg/L) | 65.70 ± 0.53* | 30.6 ± 0.69 |
| BOD (mg/L) | 14.38 ± 0.20 | 6.72 ± 0.30 |
| Electrical conductivity (salinity; µS) | 428.6 ± 0.74 | 445.7 ± 0.20 |
| Total solids (TS; mg/L) | 58.06 ± 0.26* | 41.8 ± 0.10 |
| Total suspended solids (TSS; mg/L) | 1.49 ± 0.007* | 1.23 ± 0.016 |
| Total dissolved solids (TDS; mg/L) | 56.67 ± 0.06 | 40.6 ± 0.04 |
| pH | 5.39 ± 0.008 | 5.49 ± 0.008 |
3.2. Antibiotic-Susceptible Isolates
In the current study, subculturing on MacConkey agar medium supplemented with cefotaxime or ceftazidime revealed pinkish colonies, indicating the presence of ESBL-producers for both PO and CR samples. In contrast, MacConkey agar medium supplemented with imipenem or meropenem showed growth and inhibition of CRE carriers for both PO and CR samples.
Ten resistant colonies were purified and sequenced. Data from the National Center Biotechnology Information genetic sequence database and GenBank IDs for each isolate were obtained, as shown in Table 3. ESBL and CRE susceptibility analyses revealed 10 isolates for which four species were identified in both PO and CR samples. The PO sample harbored two Klebsiella pneumoniae and two Morganella morganii strains, whereas the CR sample harbored two K. pneumoniae, two Acinetobacter pittii, and two Providencia rettgeri strains.
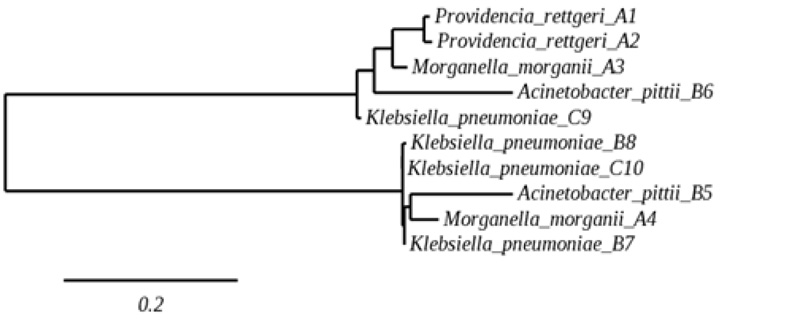
The sequences from10 isolates were used to construct a phylogenetic tree, as shown in Fig. (1). The 16S rRNA gene sequence data showed similarities among isolates where the parallel species were arranged. Susceptibility to ESBLs or carbapenems was observed for strains harboring ESBLs or for CRE.
| Site | Species | Accession Number | Resistant Gene | |||
| blaCTX-M | blaOXA | blaTEM | blaSHV | |||
| Polluted (PO) | Klebsiella pneumoniae B8 | MK024377 | + | - | - | + |
| Klebsiella pneumoniae B7 | MK024383 | - | - | - | - | |
| Morganella morganii A4 | MK024379 | + | - | - | - | |
| Morganella morganii A3 | MK024385 | - | - | - | - | |
| Clean Reference (CR) | Klebsiella pneumoniae C10 | MK024381 | - | - | - | - |
| Klebsiella pneumoniae C9 | MK024386 | - | - | - | - | |
| Acinetobacter pittii B5 | MK024378 | - | - | - | + | |
| Acinetobacter pittii B6 | MK024380 | - | - | - | - | |
| Providencia rettgeri A1 | MK024382 | - | - | - | - | |
| Providencia rettgeri A2 | MK024384 | - | - | - | - | |
3.3. Detection of β-Lactamase Genes by PCR
At a minimum, one ESBL gene in 10 isolates was distinguished by PCR (Table 3). Two isolates expressed the blaCTX gene, i.e., K. pneumoniae B8 and M. morganii A4; the blaSHV gene was detected in K. pneumoniae B8 and A. pittii B5. In contrast, no strains harbored the blaTEM and blaOXA genes. Table 1 shows the base pair (bp) sizes for the studied resistance genes, as confirmed by gel electrophoresis. The results revealed the presence of blaCTX ESBL- and CRE-resistant genes in K. pneumoniae B8 and M. morganii A4 from the PO site. Notably, blaSHV ESBL- and CRE-resistance genes were present in K. pneumoniae B8 and A. pittii B5 from PO and CR sites.
When correlating the results of antibiotic susceptibility with resistant gene screening from the PO site, we found that two isolates of M. morganii showed CRE susceptibility, and one was a carrier of the blaCTX-M gene. Thus, marine contamination by polluted effluents contributed to antibiotic-resistant bacteria. However, the isolates A. pittii and P. rettgeri were not found in the PO site.
4. DISCUSSION
The dumping of pollutants from point and diffuse source, and in cities where there is not a system of efficient sewage treatment has affected aquatic habitat. The domestic sewage is a major source of pollution, stimulating growth of bacteria and other micro-organisms, including those of fecal origin [19, 20]. The evaluation of the resistance of micro-organisms against various antimicrobials suitable for human population has been indicated to evaluate the anthropogenic contamination of a wellspring [21]. As the sewage of domestic origin is characterized by the presence of fecal material [22], it can be considered that this source of pollution most influenced the micro biota of aquatic habitat. The strains of E. coli isolated from Cascavel River have different profiles of antimicrobial resistance and were observed at the site of greater urbanization with higher prevalence of resistance. The fact that there are nearby homes to this sampling point may be a contributing factor for the identification of micro-organisms resistant to a greater number of drugs such as ampicillin, streptomycin, penicillin and tetracycline.
To enable isolation of ESBL producers, the choice of agar medium is essential, and bacterial growth on agar plates should not be considered the ultimate measurement of ESBL production [23]. Any organism that has resistance to the antibiotic present in the agar will show growth on corresponding plates [24]. The development of resistance machinery other than ESBL could be verified on selective agar. In this study, we used 1 mg/L ceftazidime and cefotaxime in order to distinguish ESBL-carrying isolates from fish fecal samples. To enable the isolation of ESBL-carrying organisms, previous studies have used a series of concentrations (0.5-2 mg/L) of cefotaxime and ceftazidime [25, 26]. Nevertheless, the ability to identify different ESBL-producing organisms using a single discerning antibiotic is unclear [25], and distinguishing among all known ESBL carriers is therefore difficult when using a single discerning medium [27].
Previous studies have revealed the high resistance of M. morganii isolates to various antibiotics, such as ceftazidime, cefepime, and cephalosporins. However, resistance to third-generation cephalosporins has arisen primarily through alterations in class A β-lactamases, including CTX-M, TEM, and SHV type enzymes [28]. Owing to the arrangement of the CTX-M gene of cefotaximase, a third-generation cephalosporin, ceftazidime resistance has been revealed, and CTX-M enzymes show greater sensitivity to cefotaxime than ceftazidime [29]. In previous studies conducted in Greece and China, the proportions of isolates resistant to meropenem and imipenem (94% and 88%, respectively) were determined in M. morganii isolates, and these β-lactams showed maximum activity towards M. morganii [30, 31].
Ceftazidime was selected for the detection of ESBL markers because this antibiotic is the most efficacious third-generation cephalosporin for most CTX-M-, SHV-, and TEM-type ESBLs [18, 32]. Among plasmid ESBL promoters, CTX-M genes often have integron cassettes or transposon fragments [33, 34]. The resistance gene blaSHV was observed in 10 isolates. In a previous study, gram-negative bacteria were found to harbor SHV-type β-lactamases [35] with glutamate and glycine replacing lysine and serine at positions 240 and 238, respectively [36]. Various studies have demonstrated the presence of more than 125 mutant SHV genes [37]. These SHV β-lactamases are the main ESBL types in the United States of America and Europe [38]. The tested M. morganii isolates harbored six blaTEM genes; the first plasmid promoters of TEM-type ESBLs were from Enterobacteriaceae, such as Proteus mirabilis, K. pneumonia, and Escherichia coli, and from non-Enterobacteriaceae, such as Pseudomonas aeruginosa [39]. In this study, we only performed genotype testing and antibiotic susceptibility analysis. Thus, further studies are needed to perform proteomics analyses and sequencing.
CONCLUSION
In conclusion, our results showed that M. morganii showed resistance to most antibiotics and harbored some β-lactamase genes. The indiscriminate use of antimicrobials exposes humans and animals to various risks, such as the selection of resistant bacteria in the aquatic environment; changes in the micro biota of growing environments and transfer of resistance to potentially pathogenic bacteria to humans [40]. These antimicrobials can be inserted into the human food chain through ingestion of water by animals in the livestock sector, contaminated fish and irrigation in agriculture, which becomes increasingly important research to assess the origin of contamination of a wellspring. Our study was limited to antibiotic susceptibility and genotype testing. Further study using sequencing technology and proteomic analysis is recommended.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Fishing was conducted according to the guidelines (Decree No. 21911, 7 November 1988) of the Ministry of Agriculture, Kingdom of Saudi Arabia. Fish sampling was authorized by the Royal Coast Guard of the Kingdom of Saudi Arabia (Decree No. 2, 3 February 1990).
HUMAN AND ANIMAL RIGHTS
All procedures performed at King Abdulaziz University abided by Royal Decree No. M/59, August 24, 2010, entitled “Research Ethics for Handling of Living Animals”.
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This project was sponsored by King Abdulaziz City for Science and Technology (KACST), Saudi Arabia (37-1948-أط)..
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


